Cell cultures
To isolate BMSCs, mice (4-week-old male, C57BL/6) had been anesthetized with isoflurane after which euthanized. The femurs and tibias had been reduce and flushed. The cells had been cultured and passaged to the second technology at 37 °C with 5% CO2.
To isolate macrophages, bone marrow cells had been handled with RBC lysis buffer (Sigma, STL, USA). The cells had been seeded for twenty-four h, and nonadherent cells had been harvested and cultured with 25 ng/mL M-CSF (Novoprotein, Jiangsu, China) for 7 days.
Bacterial strains and cultures
Porphyromonas gingivalis (P.g, ATCC 33277) was cultured in mind coronary heart infusion broth supplemented with 1 µg/mL vitamin Okay in an anaerobic tank (80% N2, 10% H2, and 10% CO2) as beforehand described [23]. Bacterial counts had been carried out through spectrophotometry (OD600 nm of 0.5 = 108 cells/mL).
Inflammatory macrophage fashions and extraction of MEVs
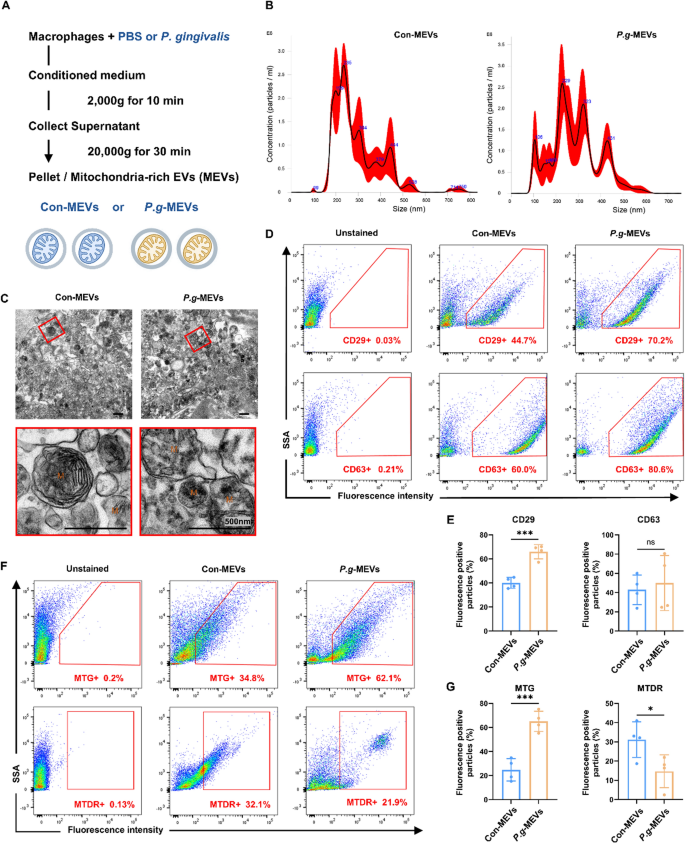
The macrophages had been contaminated with P.g for 1 h at a multiplicity of an infection (MOI) of 100 (P.g-Mφ) [24, 25]. After an infection, the cells had been washed with PBS and incubated with 0.3 mg/mL gentamycin (Sigma, STL, USA) and 6 mg/mL metronidazole (APExBio, Houston, USA) for 1 h [24, 25]. Uninfected cells had been used as controls (Con-Mφ). All tradition media and PBS had been filtered by way of a 0.1 µm filter (Millipore, Massachusetts, USA). The conditioned medium was collected, and after centrifugation at 4 ℃ [17], Con-MEVs or P.g-MEVs had been obtained (Fig. 1A). BMSCs had been incubated with MEVs (2.67 × 108/mL) for subsequent experiments.
Macrophages launch mitochondria-rich extracellular vesicles (MEVs), and inflammatory macrophages package deal extra broken mitochondria into MEVs. A Macrophage-conditioned media had been collected, and mobile particles was excluded by centrifugation at 2000×g for 10 min. Subsequent centrifugation at 20,000×g for 30 min was carried out to isolate the MEVs. MEVs from macrophages handled with PBS served as a management (Con-MEVs), and people from macrophages handled with Porphyromonas gingivalis (P.g) had been designated P.g-MEVs. B Consultant photographs of nanoparticle monitoring evaluation (NTA) for MEVs, with the horizontal axis representing diameter (nm) and the vertical axis representing particle focus (106/mL). C Consultant transmission electron microscopy (TEM) photographs of MEVs. D, E The expression of the floor markers CD29 and CD63 on MEVs was examined through circulation cytometry. F, G The contents of mitochondria (MTG) and undamaged mitochondria (MTDR) inside MEVs had been examined by circulation cytometry. Values had been imply ± SD, n = 3. *P ≤ 0.05, ***P ≤ 0.001
Quantitative RT‒PCR
Complete RNA was remoted through TRIzol. Complementary DNA (cDNA) was synthesized through the HiScript II Q RT SuperMix Equipment, in keeping with the producer’s protocol (Vazyme, Nanjing, China). Desk S1 lists the sequences of the primers used. Quantitative RT‒PCR was carried out in triplicate with a LightCycle96 utilizing FastStart Important DNA Inexperienced Grasp Combine (Roche, Basel, Switzerland). The two-ΔΔCT technique was used to investigate the outcomes.
Measurement of particle dimension and focus
The particle dimension and focus of the MEVs had been decided through a NanoSight NS300 analyzer (Malvern Panalytical, Melvern, USA). The pellets had been resuspended and detected with a 405 nm laser. Nanoparticle monitoring evaluation (NTA) was carried out with PBS as a management to find out the suitable digital camera stage and detection threshold.
Circulate cytometry evaluation
Circulate cytometry evaluation was carried out to detect the proportions of complete mitochondria and undamaged mitochondria in MEVs [26]. The particles had been stained with MitoTracker Inexperienced or MitoTracker Deep Purple (MTG or MTDR; Thermo Fisher Scientific, #M22426 or #M7514; Waltham, USA) at 37 °C for 30 min. The immunophenotype of the MEVs was characterised with CD29 and CD63 (1:200, BioLegend, #101211 and #123107; California, USA), and the particles had been labeled with antibodies for 30 min at 4 °C.
To characterize the precise floor antigens of the macrophages, the aesthetic cells had been blocked with antibodies towards CD16/32 (Elabscience, #F0997A; Wuhan, China) after which stained with antibodies towards CD11b and F4/80 (1:200, BioLegend, #102207 and #143907) at the hours of darkness for 60 min. Unstained cells had been used to regulate the voltage. FACS evaluation was carried out utilizing a BD LSRFortessa (BD, New Jersey, USA).
Transmission electron microscopy (TEM)
For TEM evaluation, pellets had been fastened in 2.5% glutaraldehyde in a single day, then with osmium tetroxide, dehydrated in ethanol, and infiltrated with acetone. Sections had been reduce through an EM UC7 ultramicrotome (Leica, Wetzlar, Germany), mounted on Formvar-coated grids, and stained with 2% uranyl acetate. Photographs had been obtained with a JEOL JEM-1200EX TEM at 80 kV [27].
Animals
Periodontitis was induced by ligation and P.g software. Orthodontic chrome steel ligature wires (0.25 mm) had been inserted from the palatal facet of the second molar of 3-month-old male C57BL/6 mice. Each 4 days, the mice had been injected with 100 μL of MEVs at a focus of two.67 × 1010/mL through tail vein (IV, intravenous injection). The mice had been randomly assigned to 5 teams: Management, Periodontitis, Periodontitis + PBS, Periodontitis + Con-MEVs, and Periodontitis + P.g-MEVs. In line with the Useful resource Equation Method, there have been 6 mice in every group [28].
Detection of mitochondrial switch in vivo
MEVs had been stained with 100 nM MitoTracker Purple at 37 °C for 30 min after which washed with PBS thrice to eradicate the dyes. The labeled MEVs (100 μL, 2.67 × 1010/mL) had been injected into the mice. At 4 h or 24 h put up injection, the mice had been euthanized, and the hearts had been perfused. The center, liver, spleen, lungs, kidneys and maxilla had been separated. Fluorescence was detected through an IVIS Spectrum system (PerkinElmer, Waltham, USA). Maxillae had been cryosectioned to evaluate mitochondrial distribution after which stained with anti-SCA1 (1:200, Abcam, #109211, Waltham, USA) and DyLight 488 secondary antibodies, with DAPI for nuclei.
Micro-computed tomography (Micro-CT)
Micro-CT scanning of the maxillae was carried out (μCT 50; SCANCO, Switzerland) with a decision of 9 µm and parameters of 70 kV and 114 mA. The DataViewer (SR Analysis, Mississauga, CA) was utilized for third-dimensional reconstruction. The space from the cementoenamel junction (CEJ) to the alveolar bone crest (ABC) was measured because the imply worth of 4 websites (distal area of the distal buccal root and palatal root of the primary molar, and mesial area of the mesial buccal root and palatal root of the second molar). The brink used for the Micro-CT knowledge analysis was 2000–10,000 Hu.
Histological staining
The maxillae had been decalcified, dehydrated, and embedded in paraffin earlier than being sectioned. They had been deparaffinized in xylene and rehydrated in graded ethanol earlier than being stained with H&E and Masson’s trichrome by staining kits (Solarbio, Beijing, China). The sections had been scanned and photographed utilizing a slide scanner (Leica Aperio AT2, Wetzlar, Germany). For immunohistochemistry (IHC) staining, the sections had been stained with an anti-LCN2 antibody or anti-RUNX2 antibody (1:200, Abcam, #318209, #192256), adopted by secondary antibodies. The imply optical density (MOD) was analyzed through a scanning microscope. A number of fields of view had been taken to calculate the typical MOD.
Mitochondrial switch coculture fashions
MEVs had been collected from macrophages labeled with MitoTracker (inexperienced). The BMSCs had been preliminarily stained with MitoTracker (purple) and Hoechst. Then, the BMSCs had been incubated with resuspended MEVs for twenty-four h, and detected with a Zeiss LSM980 Airyscan confocal microscope.
Alizarin purple S (ARS) staining
The tradition medium was eliminated, and 4% formaldehyde was added at room temperature. After being washed three extra occasions, the cells had been incubated with ARS staining answer at the hours of darkness for 30 min. Then, the formation of mineralized nodules was detected with a microscope.
Immunofluorescence
The cells had been stained with MitoTracker (inexperienced), and subsequently fastened with 4% formaldehyde. Then, the cells had been permeabilized, blocked with 3% BSA containing 0.3% Triton X-100 for 20 min, and incubated with main antibodies (LCN2, 1:200; Abcam, #318209) and secondary antibodies (Proteintech, #SA00013; Guangzhou, China). Confocal photographs had been acquired with a confocal microscope.
ATP measurement
The extent of intracellular ATP was decided with an ATP Assay Equipment (Invitrogen, California, USA). After cell lysis, the combination was harvested and centrifuged at 12,000×g for 10 min. Then, the supernatant and luminescence take a look at answer had been collected, and ATP willpower was carried out.
Western blot
Proteins had been loaded on 4–12% Bis‒Tris gels (GenScript, Piscataway, USA) and transferred to polyvinylidene fluoride membranes (Millipore, USA). After blocking with 5% BSA, the membranes had been incubated with main antibodies (LCN2, 1:1000, Abcam, #ab318209; OPA1, 1:1000, Abcam, #ab42364; OMA1, 1:1000, Affinity, #DF12435; β-actin, 1:10,000, Affinity, #AF7018, Jiangsu, China). After remedy with secondary antibodies, visualization was carried out with a ChemiDoc Western ECL system (Bio-Rad, Hercules, USA).
Proteomic evaluation of MEVs
MEVs had been remoted and dissolved in lysis buffer containing 8 M urea, 1 mM protease inhibitors, and a couple of mM EDTA. After centrifugation at 14,000×g for 20 min, the supernatant was collected and digested with trypsin. The samples had been quenched with formic acid, and LC‒MS/MS evaluation was carried out on a NanoElute UHPLC instrument and timsTOF Professional (Bruker Daltonics, Billerica, USA). The devices had been run for peptide identification and quantification through the DIA technique, through the UniProt-proteome_UP000000589_Mus_musculus.fasta database. The Gene Ontology (GO), euKaryotic Ortholog Teams (KOG), and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations and enrichment analyses had been primarily based on the UniProt database. The vital differentially expressed proteins (DEPs) with a fold change >1.5 or <0.6667 and a P-value < 0.05 had been recognized. The R Bioconductor package deal DESeq2 model 1.40.2 was used to investigate the DEPs.
LCN2-overexpressing BMSC technology
LCN2-overexpressing BMSCs had been generated through transduction with a lentiviral vector carrying the LCN2 gene (Genechem, Shanghai, China). The cDNA of LCN2 (out there at https://www.ncbi.nlm.nih.gov/nuccore/BC132071.1) was amplified from 293T cells and cloned and inserted into the Ubi-MCS-3FLAG-SV40-puromycin vector. The Ubi-MCS-3FLAG-SV40-puromycin empty vector was used as a unfavourable management (NC). BMSCs had been contaminated with a lentivirus encoding gcGFP (Ubi-MCS-3FLAG-CBh-gcGFP-IRES-puromycin) to find out the optimum MOI. BMSCs had been incubated with lentivirus and polybrene for 16 h. After tradition in contemporary medium for 72 h, optimistic cells had been chosen with puromycin. The transduction effectivity was decided by Western blotting and qRT‒PCR. The cells transfected with the LCN2 lentivirus combination had been named LCN2Hello, and the cells transfected with the management lentivirus combination had been termed NC.
Statistical evaluation
All the info had been expressed because the means ± SD. When the info had been usually distributed, an unpaired Scholar’s t-test was used for two-group comparisons, and evaluation of variance (ANOVA) was used for 3 or extra teams, adopted by Tukey’s Kramer take a look at for a number of comparisons. When the info weren’t usually distributed, the Mann–Whitney U take a look at was utilized for two-group comparisons, and the Kruskal–Wallis H take a look at was utilized for 3 or extra teams. Significance was set at a P-value < 0.05 (*P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001). Statistical analyses had been carried out with industrial software program (GraphPad Prism 9).