Excessive hnRNPA2B1 expression is related to poor prognosis and omental metastasis in OvCa sufferers
To research the existence of the sEV-miRNA sorting phenomenon in OvCa, the GSE103708 dataset was analyzed. The outcomes revealed important variations between the miRNAs recognized in OvCa cells and people detected of their corresponding sEVs (Fig. S1A-B). Proteins recognized because the potential RBPs concerned in sEV-miRNA sorting in OvCa embody members of the hnRNP household (hnRNPA2B1, hnRNPC, hnRNPG, hnRNPH1, hnRNPK, and hnRNPQ), in addition to YBX1, HuR, AGO2, IGF2BP1, MEX3C, ANXA2, ALIX, NCL, FUS, TDP-43, MVP, LIN28, SRP14, QKI, and TERT [11]. Amongst them, hnRNPA2B1, hnRNPC, hnRNPK, HuR, and FUS have been recognized as the highest 5 key molecules on the premise of the CytoHubba MCC algorithm by Cytoscape software program via the PPI community (Fig. 1A-B). Additional evaluation of the TCGA and GTEx datasets was used to find out the sensitivity and specificity of the prognosis of OvCa or regular sufferers, and the outcomes revealed that hnRNPA2B1 had the best space below the curve among the many prime 5 key molecules (Fig. 1C), indicating its potential scientific worth. Moreover, the expression of HNRNPA2B1 was greater in OvCa tissues than in regular tissues within the GSE27651 database (Fig. S1C). Excessive HNRNPA2B1 expression was considerably related to superior Worldwide Federation of Gynecology and Obstetrics (FIGO) stage of OvCa sufferers within the GSE14407 database and poor survival of OvCa sufferers within the TCGA database (Fig. S1D-E). These outcomes point out that hnRNPA2B1 could play an vital function in OvCa.
Excessive hnRNPA2B1 expression is related to poor prognosis and omental metastasis in OvCa sufferers. (A) The PPI community of RNA-binding proteins concerned within the sorting of miRNAs into small extracellular vesicles (sEVs). (B) The highest 5 important molecules recognized by the PPI community primarily based on the CytoHubba MCC algorithm by Cytoscape software program. (C) ROC curves of hnRNPA2B1, hnRNPC, hnRNPK, HuR, and FUS to find out the sensitivity and specificity of the prognosis of OvCa or regular sufferers primarily based on the TCGA and GTEx datasets. (D) Consultant IHC photographs of hnRNPA2B1 in OvCa tissues. Scale bar, 250 μm. (n = 54). (E-F) Kaplan − Meier curve of total survival (E) and development free survival (F) in OvCa sufferers with totally different hnRNPA2B1 ranges (n = 54). (G-H) Consultant IHC photographs (G) and quantification (H) of hnRNPA2B1 in OvCa tissues with or with out omental metastasis (OM). Scale bar, 50 μm. (n = 54). (I) Consultant IHC photographs of hnRNPA2B1 in tumor tissues and paired omental metastases. Scale bar, 100 μm. (J) Comparability of hnRNPA2B1 ranges between the tumor tissues and paired omental metastases (n = 19). *P < 0.05, ***P < 0.001
IHC experiments have been subsequently performed on tumor tissue chips obtained from fifty-four sufferers with OvCa to evaluate the extent of hnRNPA2B1. The affected person info is proven in Desk 1. The outcomes revealed that top hnRNPA2B1 ranges have been strongly correlated with superior FIGO stage and omental metastasis (Fig. 1D; Desk 1). Kaplan-Meier survival evaluation of the fifty-four sufferers with OvCa revealed that OvCa sufferers with excessive hnRNPA2B1 ranges had considerably shorter total survival (OS) and progression-free survival (PFS) than these with low hnRNPA2B1 ranges (Fig. 1E-F). As proven in Fig. 1G-H, a better degree of hnRNPA2B1 in tumor tissues was noticed in sufferers with omental metastasis than in sufferers with out metastasis. Moreover, a complete of 19 major tumors, together with their matched omental metastases, have been utilized in IHC experiments, and the outcomes revealed that the expression of hnRNPA2B1 in tumor tissue was decrease than that in paired omental metastases (Fig. 1I-J). These findings counsel that hnRNPA2B1 may be associated to the omental metastasis of OvCa.
Knockdown of hnRNPA2B1 suppresses tumor metastasis in vivo
Additional research have been performed to discover the function of hnRNPA2B1 in selling metastasis in vivo. First, we assessed the extent of hnRNPA2B1 in OvCa cell traces, and we discovered that hnRNPA2B1 expression was upregulated within the ES-2 extremely metastatic cells (ES-2-HM cells) in contrast with that in ES-2 cells (Fig. S2A-C). We then used ES-2-HM and OVCAR-4 cells to knock down hnRNPA2B1 by way of the CRISPR-Cas9 method (Fig. S2D-E). Moreover, luciferase-labeled ES-2-HMsgNC or ES-2-HMsghnRNPA2B1 cells have been used to assemble spontaneous orthotopic tumor fashions in nude mice, and the bioluminescence sign was dynamically monitored via in vivo imaging (Fig. 2A). The outcomes revealed that the bioluminescence sign density within the sghnRNPA2B1 group decreased quickly in contrast with that within the sgNC group (Fig. 2B). The sign change in every mouse’s fluorescence sign was proven in Fig. S2F. Ex vivo bioluminescence detection revealed that the bioluminescence depth of belly organs and omental tissues within the sgNC group was larger than that within the sghnRNPA2B1 group (Fig. 2C-D). H&E staining of omental tissues revealed omental metastasis within the sgNC group, whereas inflammatory infiltration was noticed within the sghnRNPA2B1 group (Fig. 2E). Utilizing GEPIA2 (TCGA datasets), our research recognized important constructive correlations between hnRNPA2B1 and the gelatinases MMP2 and MMP9 in OvCa (Fig. S2G). The degrees of MMP2, MMP9, Ki67, and hnRNPA2B1 in tumor tissues from nude mice have been considerably decrease within the sghnRNPA2B1 group than within the sgNC group (Fig. 2F-H). In conclusion, these findings point out that hnRNPA2B1 might enhance the colonization capability of tumor in omental metastases.
Knockdown of hnRNPA2B1 suppresses tumor metastasis in vivo. (A) Consultant bioluminescence photographs of orthotopic ovarian xenografts generated from luciferase-labeled ES-2-HM cells with (n = 4) or with out (n = 4) hnRNPA2B1 knockdown in vivo (above), and from belly organs remoted ex vivo (under). L for liver, Okay for kidney, M for mesentery, P for pancreas, O for omentum, S for spleen, T for tumor, and F for fats pad. (B) The overall fluorescence depth at totally different remark factors. (C-D) Scatter plot displaying the bioluminescence alerts of all belly organs (C) and omentum (D) ex vivo. (E) Consultant H&E photographs of omental tissues from the nude mice described above. Scale bar, 100 μm. (F-H) Consultant H&E photographs, IHC (F) photographs and quantification of hnRNPA2B1, MMP2, MMP9 (G), and Ki67 (H) in tumors of mice. Scale bar, 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001
Tumoral hnRNPA2B1 is positively correlated with omental myofibroblasts
To evaluate which of essentially the most important cells within the TME promote OvCa omental metastasis, the GSE147082 dataset was used to guage the distribution of the subpopulations of omental tissues from OvCa sufferers on the single-cell degree. The outcomes revealed that fibroblasts composed the vast majority of the subpopulations (Fig. 3A, Fig. S3A-D). Amongst these fibroblast clusters, myCAFs constituted the best proportion of cells (Fig. 3B-C). Gene set variation evaluation (GSVA) evaluation of GSE193875 and GSE147082 indicated that the myCAFs was extra widespread in omental metastases than in non-omental metastases (Fig. 3D). The marker genes related to myCAFs have been recognized as ACTA2, FAP, and POSTN (Fig. 3E, Fig. S3E-H). These outcomes point out that myCAFs could play an vital function within the OvCa omental metastasis.
Tumoral hnRNPA2B1 expression is positively correlated with omental myofibroblasts. (A) UMAP plot displaying the distinct cell sorts recognized by marker genes from OvCa sufferers within the GSE147082 dataset. (B) UMAP plot of cancer-associated fibroblast (CAFs) displaying seven clusters. (C) Bar plots displaying the proportions of every CAFs cluster in each pattern. (D) Heatmap displaying GSVA scores for every CAFs cluster (GSE147082) in OvCa sufferers (GSE193875) with or with out omental metastasis (OM). (E) UMAP plot displaying the expression of ACTA2, FAP, and POSTN in all CAFs clusters. (F) Consultant morphology of #1357 omental-derived major fibroblasts (ODPFs) (with out omental metastasis) and #1332 ODPFs (with omental metastasis) decided by bright-field microscopy (left) and after F-actin staining (proper). Scale bar, 200 μm (left). Scale bar, 100 μm (center panel). Scale bar, 50 μm (proper). (G) qRT–PCR evaluation of ACTA2, FAP, and POSTN expression in ODPFs. (H) Western blot evaluation of the α-SMA, FAP, and periostin degree in ODPFs. (I-Okay) Consultant IHC photographs and quantification of α-SMA and picrosirius purple staining in tumor tissues and omental tissues from orthotopic xenografts generated from ES-2-HMsgNC or ES-2-HMsghnRNPA2B1 cells. Scale bar, 100 μm. (L) Consultant IHC photographs of hnRNPA2B1 in ovarian lesions, and displaying the expression of α-SMA or periostin in corresponding paired omental tissues. Scale bar, 100 μm. (M-N) Scatter plot displaying the correlation between hnRNPA2B1 ranges in ovarian lesions and α-SMA or periostin ranges in paired omental tissues. *P < 0.05, **P < 0.01, ***P < 0.001
We subsequently extracted the ODPFs from the omental tissues of OvCa sufferers. Underneath gentle microscopy and immunofluorescence staining, in contrast with ODPFs with out metastasis, ODPFs derived from metastatic omental tissues introduced enlarged cell our bodies and elongated cytoskeletal buildings (Fig. 3F). The myCAFs markers α-SMA, FAP, and periostin, have been extra plentiful in ODPFs remoted from omental metastases than in ODPFs remoted from non-omental metastases, as revealed by qRT-PCR and Western blotting assays (Fig. 3G-H, Fig. S4A). Moreover, tumor tissues and omental tissues have been remoted from spontaneous orthotopic tumor fashions mice by way of ES-2-HMsgNC or ES-2-HMsgHNRNPA2B1 cells. Then, picrosirius purple staining was used to detect collagen deposition within the stroma, and α-SMA staining was used to detect myCAF activation. The outcomes revealed a big discount within the degree of α-SMA and within the space of picrosirius purple staining in tumor tissues and omental tissues from the sghnRNPA2B1 group in contrast with these from the sgNC group, suggesting that hnRNPA2B1 promotes collagen deposition and myofibroblast activation (Fig. 3I-Okay). Moreover, tumoral hnRNPA2B1 expression was positively correlated with the degrees of α-SMA and periostin in paired omental tissues from OvCa sufferers (Fig. 3L-N, Desk S5). These findings counsel that tumoral hnRNPA2B1 may be concerned in omental myCAF activation in OvCa.
Tumoral hnRNPA2B1 facilitates omental metastasis and myofibroblast activation by way of small extracellular vesicles
The sEVs derived from OvCa tumor cells have been purified from the conditioned medium utilizing the differential ultracentrifugation methodology, as described beforehand [4]. Underneath a transmission electron microscope, the sEVs exhibited a saucer-shaped double-membrane construction with melancholy on one aspect (Fig. 4A). Nanoparticle monitoring evaluation was used to measure the dimensions and variety of vesicles. The outcomes revealed that the height focus diameter of sEVs derived from OVCAR-4sgNC cells was 136.1 nm, whereas for sEVs derived from OVCAR-4sghnRNPA2B1 cells, the height focus diameter was 137.4 nm. The proportion of sEV particles starting from 50 to 200 nm was 80.99% for OVCAR-4 sgNC cells and 78.59% for OVCAR-4sg − hnRNPA2B1 cells. These outcomes confirmed the profitable isolation of sEVs (Fig. 4B). Among the many markers, sEVs expressed the constructive markers CD63, CD81, and Tsg101 however not the detrimental marker calnexin (Fig. 4C). ODPFs have been subsequently handled with PBS, sEVsgNC, or sEVsghnRNPA2B1. Western blotting revealed that the degrees of the myCAF markers α-SMA, FAP, and periostin have been considerably elevated in ODPFs handled with sEVsgNC in contrast with these in ODPFs handled with the detrimental management (PBS) or sEVsghnRNPA2B1 (Fig. 4D, Fig. S4B-C). To research whether or not hnRNPA2B1 is current within the vesicle lumen of sEVs, dot blot assays have been carried out. The outcomes revealed that CD63 was detectable on the floor of sEVs and was additionally current within the vesicle lumen, whereas hnRNPA2B1 was detected solely below detergent-containing situations (Fig. S5A). Functionally, Transwell assays demonstrated that sEVsgNC notably enhanced the migration and invasion talents of ODPFs in contrast with these of PBS or sEVsghnRNPA2B1 (Fig. 4E-G, Fig. S5B). Morphologically, ODPFs handled with sEVsgNC had enlarged cell our bodies and an prolonged spindle look in contrast with these handled with PBS or sEVsghnRNPA2B1 (Fig. S5C). Collectively, these information counsel that tumoral hnRNPA2B1 promotes myCAF activation by way of sEVs.
Tumoral hnRNPA2B1 promotes omental metastasis and myofibroblast activation via small extracellular vesicles. (A) Consultant transmission electron microscopy micrographs of small extracellular vesicles (sEVs) secreted by OVCAR-4 cells. Scale bar 100 nm. (B) Nanoparticle monitoring evaluation of sEVs secreted by OVCAR-4 cells. (C) Western blotting evaluation of constructive sEV markers (CD63, CD81, and Tsg101) and the detrimental marker Calnexin in whole OvCa cells lysates (TCL) and the corresponding sEVs. (D) Western blot evaluation of the α-SMA, FAP, and periostin ranges in #1357 (left) ODPFs and #1368 (proper) ODPFs handled with sEVs derived from OvCa cells. (E) Consultant photographs of Transwell assays in #1370 ODPFs handled with PBS or the indicated sEVs derived from OVCAR-4 cells. Scale bar, 200 μm. (F-G) Quantification of Transwell assays for ODPFs handled with PBS or the indicated sEVs derived from OvCa cells. (H) Consultant bioluminescence photographs of belly metastases in vivo (left) and belly organs remoted ex vivo after sacrifice (proper) within the indicated teams. (n = 4 per group) (I) Bioluminescence alerts at totally different remark factors. (J) Scatter plot displaying the bioluminescence sign of the omentum ex vivo. (Okay-L) Consultant IHC photographs (Okay) and quantification (L) of α-SMA and periostin in tumor tissues and omental tissues from nude mice. Scale bar, 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001
Subsequently, we explored whether or not the sEVs derived from tumoral hnRNPA2B1 promote OvCa omental metastasis in vivo. Spontaneous orthotopic tumor fashions in nude mice have been established, after which the mice have been intraperitoneally injected with PBS, sEVsgNC, or sEVsghnRNPA2B1 (Fig. 4H). In contrast with these within the sEVsghnRNPA2B1 or PBS group, the in vivo bioluminescence alerts within the sEVsgNC group exhibited a speedy enhance (Fig. 4I). Ex vivo bioluminescence demonstrated that the bioluminescence alerts in omental tissues have been larger within the sEVsgNC group than the sEVsghnRNPA2B1 or PBS teams (Fig. 4J). Moreover, we found that the degrees of myCAF markers, as indicated by α-SMA and periostin, have been larger within the sEVsgNC group than in these from the sEVsghnRNPA2B1 or PBS teams (Fig. 4Okay-L). These outcomes point out that tumoral hnRNPA2B1 enhances the power of sEVs to advertise myCAF activation and OvCa omental metastasis.
Tumoral hnRNPA2B1 mediates miRNA sorting to small extracellular vesicles in a UAG sequence-dependent method
Given the sorting perform of hnRNPA2B1 in sEVs, the amount and cargoes of sEVsgNC and sEVsghnRNPA2B1 have been analyzed. The particle counts, protein ranges, and RNA ranges of sEVs derived from equal numbers of cells have been measured, and the outcomes revealed no important variations between sEVsgNC and sEVsghnRNPA2B1 (Fig. 5A-C). Moreover, sEV uptake assays have been carried out, and the outcomes revealed that each sEVsgNC and sEVsghnRNPA2B1 have been taken up by ODPFs with out notable variations (Fig. 5D). To display the important miRNAs sorted by hnRNPA2B1 in sEVs, high-throughput miRNA sequencing was carried out on the sEVsgNC and sEVsghnRNPA2B1 derived from OVCAR-4 cells. As proven in Fig. 5E, in contrast with these in sEVsgNC, there have been eleven considerably downregulated miRNAs and ten considerably upregulated miRNAs in sEVsghnRNPA2B1. Among the many eleven downregulated miRNAs, the six miRNAs (miR-21-5p, miR-224-5p, miR-92a-3p, miR-93-5p, miR-29a-3p, and miR-125a-5p) have been enriched within the OVCAR-4 cells. To determine whether or not these miRNAs are transported from cells to sEVs, we in contrast the degrees of miRNAs in each OvCa cells and their corresponding sEVs (Fig. 5F-G, Fig. S6A). The outcomes revealed important will increase within the sEV/cells ratios of miR-21-5p, miR-29a-3p, and miR-224-5p upon hnRNPA2B1 knockdown (Fig. 5H-I). To verify that these miRNAs have been contained throughout the sEVs, the sEVs have been handled with RNase within the absence or presence of Triton X-100. The outcomes revealed that the three extremely enriched miRNAs have been immune to RNase within the absence of Triton X-100 (Fig. S6B). To discover the sorting capability of hnRNPA2B1 in vivo, we utilized sEVs derived from mice subcutaneous tumor tissues (sEVTissue) and the ascites of OvCa sufferers for validation. The sEVTissue ranges of miR-21-5p, miR-29a-3p, and miR-224-5p within the sghnRNPA2B1 group have been considerably decrease than these within the sgNC group, whereas the entire RNA ranges of sEVTissue weren’t considerably totally different. (Fig. 5J-Okay). Moreover, the bottom ranges of miR-29a-3p and miR-224-5p in ODPFs handled with ascites-derived sEVs have been coincident with the bottom degree of hnRNPA2B1 in ascites-derived sEVs from the Affected person #1094 in contrast with Affected person #1121 and Affected person #1296 (Fig. S6C-D, Desk S5). Collectively, these outcomes point out that these miRNAs actively combine into sEVs by way of a course of regulated by hnRNPA2B1 in OvCa cells.
Tumoral hnRNPA2B1 mediates miRNAs sorting to small extracellular vesicles within the UAG sequence-dependent method. (A-C) The particle counts (A), the protein concentrations (B) and the RNA ranges (C) in whole small extracellular vesicles (sEVs) from equal quantities of cells have been measured in OVCAR-4 cells. (D) Consultant photographs of #1357 ODPFs uptake PKH67-labeled sEVs derived from OVCAR-4 cells. Purple staining represents F-actin, inexperienced staining represents the PKH67-labeled sEVs, and blue staining represents DAPI. Scale bar, 100 μm (E) Hierarchical clustering evaluation of miRNA expression profiles in sEVs derived from OVCAR-4 cells after hnRNPA2B1 downregulation. (F-G) Differential ranges of six miRNAs in OVCAR-4 cells (F) and in sEVs (G) derived from OVCAR-4 cells with or with out hnRNPA2B1 knockdown. (H-I) The sEV/cell ratio after hnRNPA2B1 knockdown of miRNAs between OvCa cells and sEVs in OVCAR-4 cells (H) or ES-2-HM cells (I). (J) The RNA quantitative evaluation of whole sEVs derived from subcutaneous tumor tissues. (Okay) qRT–PCR evaluation of the extent of the miRNAs in sEV derived from subcutaneous tumor tissues. (L) RIP assays displaying that hnRNPA2B1 instantly interacted with the miRNAs relative to IgG. (M) Western blot evaluation of hnRNPA2B1 ranges in samples derived from miRNA pulldowns carried out in OVCAR-4 cells. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not important
To look at the direct interplay between hnRNPA2B1 and these miRNAs, RIP assays and miRNA pull-down assays have been carried out. In contrast with these within the IgG group, the teams handled with the hnRNPA2B1 antibody have been considerably enriched (Fig. 5L). Additional sequence evaluation revealed that miR-21-5p, miR-29a-3p, and miR-224-5p all comprise the UAG motif, a sequence that instantly combines with hnRNPA2B1 [20] (Desk S6). Wild-type and mutated miRNAs have been labeled with biotin after which transfected into OvCa cells, adopted by immunoprecipitation to detect hnRNPA2B1 protein ranges. The outcomes indicated that wild-type biotin-miRNAs work together with hnRNPA2B1, whereas the binding capability was decreased when the UAG motif within the miRNAs was mutated (Fig. 5M). Furthermore, dot-blot assays confirmed the presence of hnRNPA2B1 within the sEVsgNC whereas revealing a decreased degree of hnRNPA2B1 within the sEVsghnRNPA2B1 derived from OvCa cells (Fig. S6E). These findings counsel that tumoral hnRNPA2B1 instantly binds to the UAG motif in a sequence-dependent method to help within the sorting of miRNAs into sEVs.
Tumoral hnRNPA2B1 triggers myofibroblast activation via miRNAs enriched in small extracellular vesicles
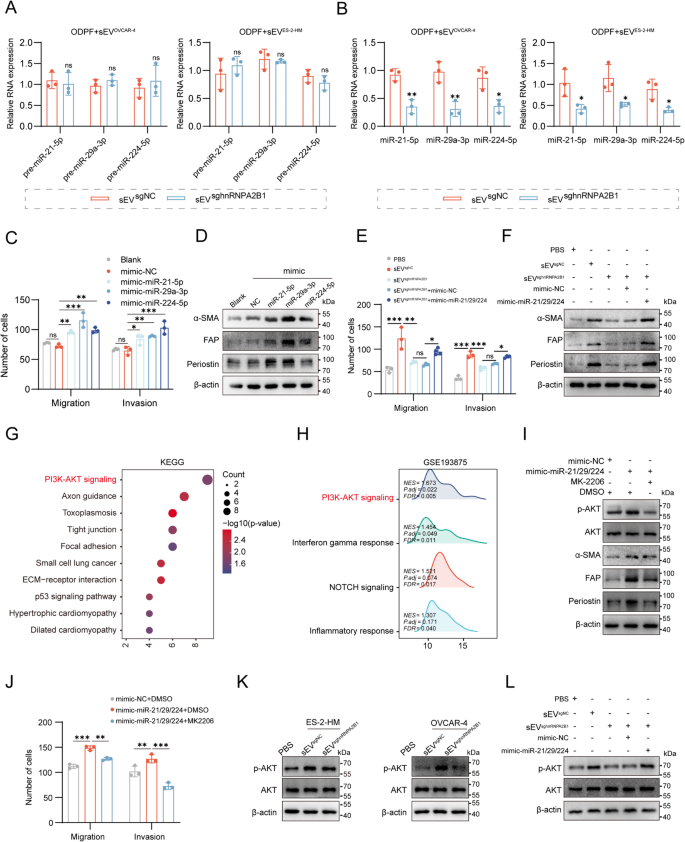
To verify whether or not sEV-miRNAs activate myCAFs, ODPFs have been handled with sEVsgNC or sEVsghnRNPA2B1. In contrast with these in ODPFs handled with sEVsgNC, the mature miRNA ranges have been decrease within the ODPFs handled with sEVsghnRNPA2B1, whereas the degrees of precursor miRNAs weren’t considerably totally different among the many totally different teams (Fig. 6A-B). To elucidate the roles of miR-21-5p, miR-29a-3p, and miR-224-5p (miR-21/229/224) in myCAF activation, ODPFs have been transfected with mimics of those miRNAs (Fig. S7A). Transwell assays and Western blotting have been subsequently carried out to discover the phenotypic modulation of myCAF activation, which was characterised by improved migration and invasion capacities in addition to elevated ranges of α-SMA, FAP, and periostin. The outcomes confirmed that the upregulation of those miRNAs promoted myofibroblast activation (Fig. 6C-D, Fig. S7B, Fig. S9A). And up-regulation of miR-21-5p, miR-29a-3p, and miR-224-5p led to decrease expression of PTEN (Fig. S7C). To find out whether or not miRNAs activation in myCAFs was induced by hnRNPA2B1-associated sEVs, ODPFs have been handled with PBS, sEVsgNC, sEVsghnRNPA2B1, sEVsghnRNPA2B1 mixed with miR-21/229/224 mimics, or sEVsghnRNPA2B1 mixed with miRNA-NC mimics. We discovered that, in contrast with sEVsgNC, sEVsghnRNPA2B1 largely decreased ODPF migration and invasion, together with exhibiting low ranges of α-SMA, FAP, and periostin. Moreover, the upregulation of those miRNAs reversed the suppressive impact of sEVsghnRNPA2B1 on myCAFs (Fig. 6E-F, Fig. S7D, Fig. S9B). These findings point out that sEV-miRNAs activate myCAFs, that are regulated by hnRNPA2B1 in tumors.
Tumoral hnRNPA2B1 induces myofibroblast activation via miRNAs enriched in small extracellular vesicles. (A-B) qRT-PCR evaluation of the degrees of pre-miRNAs and mature miRNAs in ODPFs handled with sEVsgNC or sEVsghnRNPA2B1. (C) Quantification of Transwell assays in ODPFs after the overexpression of three miRNAs. (D) Western blot evaluation of the α-SMA, FAP, and periostin ranges in #1371 ODPFs after the overexpression of the three miRNAs. (E) Quantification of ODPFs Transwell assays within the indicated teams. (F) Western blot evaluation of α-SMA, FAP, and periostin ranges in #1488 ODPFs within the above teams. (G) KEGG pathway enrichment evaluation of genes focused by miR-21-5p, miR-29a-3p, and miR-224-5p. (H) GSEA of CAFs with or with out omental metastases within the GSE193875. (I) Western blot evaluation of the α-SMA, FAP, periostin, AKT, and phosphorylated AKT (p-AKT) degree in #1505 ODPFs within the specified group. (J) Quantification of ODPFs Transwell assays within the talked about group. (Okay) Western blot evaluation of AKT and p-AKT ranges in #1404 ODPFs (left) or #1426 ODPFs (proper) cocultured with sEVsgNC or sEVsghnRNPA2B1 derived from ES-2-HM (left) or OVCAR-4 cells (proper). (L) Western blot evaluation of AKT and p-AKT ranges in #1395 ODPFs within the indicated teams. *P < 0.05, **P < 0.01, ***P < 0.001; ns, not important
To guage the pathways concerned in myCAF activation, Kyoto Encyclopedia of Genes and Genomes (KEGG) evaluation was carried out on the goal genes of the three miRNAs, and the outcomes confirmed that the PI3K/AKT signaling pathway was considerably enriched (Fig. 6G). Gene set enrichment evaluation (GSEA) of the GSE193875 dataset additionally revealed that the PI3K/AKT pathway was enriched in CAFs remoted from the OvCa sufferers with omental metastasis in contrast with that in CAFs with out metastasis (Fig. 6H). Then, ODPFs have been transfected with the miRNA mimics and uncovered to MK2206 (an AKT inhibitor). In contrast with the detrimental management, the miRNA mimics improved the α-SMA, FAP, and periostin ranges, and elevated the migratory and invasion capacities of ODPFs. Furthermore, MK2206 largely inhibited PI3K/AKT pathway activation and partially reversed the myCAF activation induced by miRNA mimics (Fig. 6I-J, Fig. S8A, Fig. S9C). To evaluate the activation of the pathways induced by hnRNPA2B1-mediated sEVs, Western blotting assays was carried out, and the outcomes confirmed that sEVsgNC activated the pathway in ODPFs in contrast with that in PBS or sEVsghnRNPA2B1 (Fig. 6Okay, Fig. S9D). Then, ODPFs have been uncovered to PBS, sEVsgNC, or sEVsgNC mixed with MK2206, and Western blotting assays have been carried out. The outcomes confirmed that the PI3K/AKT inhibitor (MK2206) reversed the sEVsgNC-induced activation of the PI3K/AKT pathway (Fig. S7B-E). As well as, the degrees of the myCAF markers have been considerably decreased in ODPFs handled with sEVsgNC mixed with MK2206 in contrast with these in ODPFs handled with sEVsgNC (Fig. S7F-I). Functionally, Transwell assays demonstrated that MK2206 notably rescued the improved the migration and invasion talents of ODPFs induced by sEVsgNC (Fig. S7J-M). These findings strongly help the function of the PI3K/AKT pathway in tumoral hnRNPA2B1-mediated myCAF activation. To discover whether or not sEV-miRNAs affected by hnRNPA2B1 set off this pathway, ODPFs have been handled with PBS, sEVsgNC, sEVsghnRNPA2B1, sEVsghnRNPA2B1 mixed with miR-21/229/224 mimics, or sEVsghnRNPA2B1 mixed with miR-NC mimics. We discovered that the upregulation of miRNAs counteracted the inhibition of the PI3K/AKT pathway induced by sEVsghnRNPA2B1 (Fig. 6L, Fig. S9E). These outcomes point out that sEVs set off the PI3K/AKT pathway to activate myCAFs by way of a course of mediated by hnRNPA2B1 in OvCa cells.
Tumoral hnRNPA2B1 prompts myofibroblasts to advertise tumor metastasis
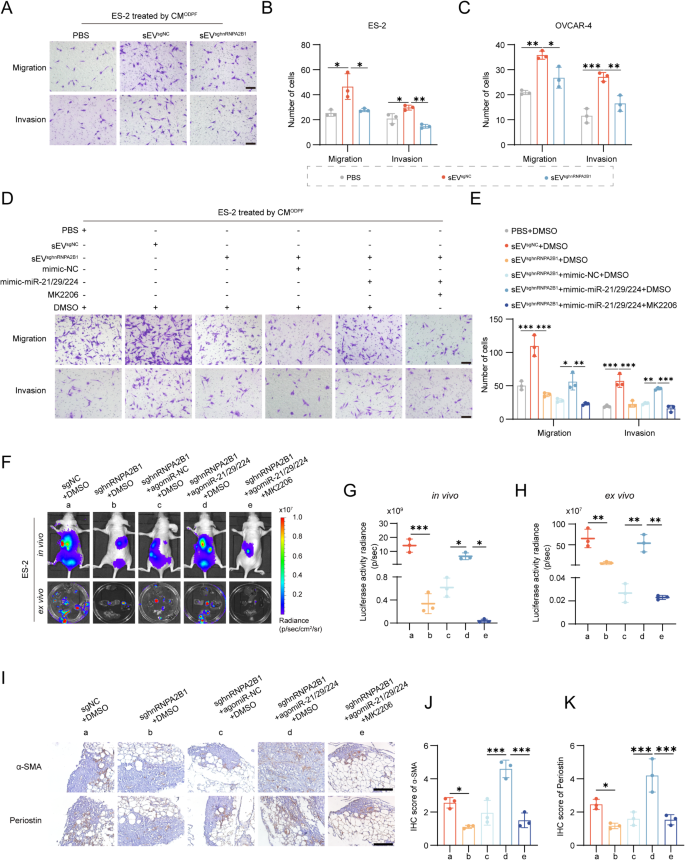
To find out the impression of sEV-activated myCAFs on OvCa cells, we handled OvCa cells with conditioned medium (CM) from ODPFs which handled with PBS, sEVsgNC, or sEVsghnRNPA2B1. Transwell assays confirmed that sEVsgNC group considerably promoted the migration and invasion of OvCa cells in contrast with PBS, and sEVsghnRNPA2B1 inhibited these talents as compared with sEVsgNC (Fig. 7A-C, Fig. S8N). To additional decide the function of myCAFs activated by sEV-miRNAs in OvCa cells, ODPFs have been pre-cocultured with PBS, sEVsgNC, or sEVsghnRNPA2B1, and ODPFs within the sEVsghnRNPA2B1 group have been transfected with miRNA mimics, adopted by incubation with MK2206. The CM collected from the ODPFs of varied teams was utilized for treating OvCa cells. Transwell assays confirmed that miRNA mimics partially reversed the decreased invasion and migration capabilities of OvCa cells induced by sEVsghnRNPA2B1, and MK2206 attenuated the improved migration and invasion talents of OvCa cells improved by the miRNA mimics (Fig. 7D-E). To validate the metastasis-promoting function of myCAFs activated by tumoral hnRNPA2B1 in vivo, nude mice have been intraperitoneally injected with sgNC or sghnRNPA2B1 cells, and mice injected with sghnRNPA2B1 cells have been handled with agomiR-21/29/224 and MK2206 (Fig. 7F). In vivo bioluminescence imaging indicated that the belly alerts within the sghnRNPA2B1 group have been considerably decrease than these within the sgNC group. Furthermore, agomiR-21/29/224 reversed the detrimental results of sghnRNPA2B1, and MK2206 notably suppressed these results (Fig. 7G). Furthermore, the development of the bioluminescence alerts in vivo was mirrored by the bioluminescence depth of the belly organ ex vivo (Fig. 7H). Equally, our findings revealed that sghnRNPA2B1 decreased the degrees of α-SMA and periostin within the omental tissues of mice in contrast with sgNC. Furthermore, agomiR-21/29/224 successfully restored the decreased ranges of α-SMA and periostin in sghnRNPA2B1 cells. Moreover, therapy with MK2206 mitigated the rise within the ranges of α-SMA and periostin that have been attenuated by agomiR-21/29/224 (Fig. 7I-Okay). In abstract, these outcomes collectively demonstrated that tumoral hnRNPA2B1 prompts myCAF by way of the PI3K/AKT pathway to advertise tumor metastasis.
Tumoral hnRNPA2B1 prompts myofibroblasts to advertise tumor metastasis. (A) Consultant photographs of Transwell assays of ES-2 cells uncovered to conditioned medium (CM) from ODPFs precocultured with PBS, sEVsgNC, or sEVsghnRNPA2B1 derived from OvCa cells. Scale bar, 100 μm. (B-C) Histogram evaluation of Transwell assays of ODPFs in ES-2 or OVCAR-4 cells within the talked about teams. (D-E) Consultant photographs and quantification of Transwell assays of ES-2 cells uncovered to the CM from ODPFs in indicated teams. Scale bar 100 μm. (F) Consultant bioluminescence photographs of belly metastases in vivo (above) and belly organs remoted after sacrifice ex vivo (under) within the indicated teams (n = 3 per group). (G-H) Scatter plot displaying the bioluminescence sign in vivo (G) and the bioluminescence sign of all belly organs ex vivo (H). (I-Okay) Consultant IHC photographs (I) and quantification of α-SMA (J) and periostin (Okay) in omental tissues from nude mice. Scale bar, 100 μm. *P < 0.05, **P < 0.01, ***P < 0.001