Culturing cells
The HEK293F (Thermo Fisher Scientific) and HMEC-1 (ATCC-3243) cell strains have been used on this examine. The medium used to domesticate the HEK293F cells was Freestyle 293 medium (Thermo Fisher) with 100 µg/mL penicillin-streptomycin answer (Cytiva Hyclone). The HEK293F cells have been cultured each adherently and in suspension. When cultured in suspension they have been cultured in a shaking incubator with shaking at 130 rpm at 37 °C, 70% humidity, and 5% CO2. When cultured as adherent cultures, 10% FBS (Cytiva Hyclone) was added to the medium and the cells have been cultured at 37 °C with 5% CO2 in a daily incubator. The HMEC-1 cell line was cultured adherently in MCDB131 medium (with out l-glutamine, ThermoFisher Scientific) supplemented with 10 ng/mL human EGF recombinant protein (ThermoFisher Scientific), 1 µg/mL hydrocortisone-water soluble (Merck), 10 mM glutamine (Cytiva Hyclone), 100 µg/mL penicillin-streptomycin answer (Cytiva Hyclone), and 10% FBS (Cytiva Hyclone).
Growing the steady LFA-1 clone
The steady LFA-1 clone was developed through sequential transfection of wild-type (WT) HEK293F cells with human CD18 and human CD11a individually. Plasmids with the sequences for CD11a and CD18 for the transfections have been bought from GenScript (Supplementary Fig. 1A and 1B). For the transfection with CD18 the cells have been grown in suspension, and for the transfection with CD11a the cells have been rising adherently. The transfection with CD18 of cells rising in suspension was carried out utilizing the FectoPro system (Polyplus) in accordance with the producer’s protocol. Briefly, on the day of transfection the cultures have been adjusted to a focus of two × 106 cells/mL in Freestyle 293 medium. For the transfection, IMDM media (Lonza) was used because the dilution medium. In a sterile tube, the IMDM (0.1 mL/mL remaining quantity), plasmid (0.8 µg/mL remaining quantity), and transfection reagent (0.8 µL/mL remaining quantity) have been mixed. As a adverse management, a mock transfection was carried out utilizing IMDM alone with out plasmid. After a 20 min incubation at room temperature, the combination was added to the cells and the cells have been positioned in a 37 °C incubator.
The transfection of adherently rising CD18 clones with CD11a was carried out with the Lipofectamine 2000 system (Thermo Fisher) in accordance with the producer’s protocol. Briefly, two days previous to the transfection 1.0 × 106 cells have been seeded on a 6-well plate (Falcon) in 2 mL development medium (Freestyle 293 medium with 10% FBS). On the day of transfection, the wells have been roughly 90–95% confluent, and the expansion medium within the wells was changed with 1.5 mL recent development medium. A mix of 4 µg of plasmid with 10 µL of the Lipofectamine 2000 transfection reagent in a complete quantity of 500 µL IMDM medium (Cytiva Hyclone) was allowed to incubate for 20 min at room temperature. Following this incubation, the combination was gently blended with the expansion medium within the wells. The identical combination with out the plasmid was added to the cells for the mock transfection.
Producing a pure clone
For choice, the antibiotics G418 (500 µg/mL) and Hygro B (125 µg/mL) have been added 48 h after transfection. The expansion medium was modified each 48 h, and swimming pools of clones have been expanded or cut up as required. When the viability of the mock tradition started to say no, a range process was used to generate pure clones. Each the transfected and mock teams underwent similar choice processes involving seeding out the transfected cells in a sequence of 10× dilutions on Petri dishes. After round two weeks, 30–50 clones have been picked from the Petri dishes utilizing a pipette by gently scraping the floor of the petri dish after which accumulating 50 µL of the media with the cells. The picked cells have been added to a 96-well plate (Falcon) already containing 150 µL media for a complete of 200 µL media per nicely. The clones have been expanded as wanted and analyzed through circulate cytometry to establish the clones with the best expression of the specified membrane proteins.
Movement cytometry
To find out the effectivity of the transfections and the expression of the specified membrane proteins, the cells have been analyzed by circulate cytometry on both a BD FACS Aria II cell sorter or a BD FACSVerse circulate cytometer working BD FACSSuite software program (BD Biosciences). One milliliter of ice-cold FACS buffer (PBS with 1% FBS) was added to 100,000 cells, which have been then pelleted at 200 × g for 10 min at 4 °C. The cells have been then resuspended in 50 µL of human IgG (1 mg/mL in D-PBS) and incubated for 15 min at 4 °C. After this incubation, antibodies particular for the membrane proteins of curiosity have been then added along with a viability dye for 30 min at 4 °C. The next antibodies and viability dyes have been used for the staining step: BD Pharmingen FITC mouse anti-human CD18 (Clone 6.7, BD Biosciences), BD Pharmingen PE mouse anti-human CD11a (Clone HI111, BD Biosciences), BD Pharmingen 7-AAD (BD Biosciences), and Invitrogen LIVE/DEAD Fixable Aqua Lifeless cell stain package (Thermo Fisher). The cells have been washed with 2 mL FACS buffer after which resuspended in a remaining quantity of 350 µL of FACS buffer. A complete of 10,000 occasions have been collected for every pattern, and the information have been analyzed utilizing FlowJo software program (Tree Star Inc, Ashland, OR, USA).
Binding assay of LFA-1 cells with ICAM-1 coating
Wells in a 96-well strip plate (Thermo Fisher Scientific, Black microplate F96) have been coated with ICAM-1 by introducing a 100 µL answer containing 5 µg/mL of the ligand in a coating buffer (0.1 M NaHCO3, pH 9.6) to the wells. Utilizing the identical focus and process, wells have been additionally coated with anti-CD18 (MEM-48, Thermo Fisher Scientific), bovine serum albumin, or human IgG as controls. Unfavorable controls consisted of wells receiving solely the coating buffer. The plates have been subsequently sealed with parafilm and incubated in a single day at 4 °C. Following incubation, the plates have been inverted, and the options have been faraway from the wells, which have been then washed twice with 150 µL of washing buffer (0.1% Tween in PBS). Subsequently, the wells have been blocked with 150 µL of blocking buffer (1% BSA in PBS) per nicely at room temperature for 1 h, concurrent with cell labelling. Cell labelling was carried out utilizing the fluorescent dye PKH67 (Sigma-Aldrich) following the producer’s protocol. Briefly, the required variety of cells was pelleted by centrifugation at 200 × g for 10 min, resuspended in 1 mL diluent C, and blended with a 2× dye working answer composed of 1 mL diluent C and 4 µL PKH67. After an incubation interval of two–5 min, the response was halted by including 2 mL of 1% BSA and incubating for 1 min at nighttime. Extra dye was eliminated by washing the cells twice with PBS.
The blocking buffer was faraway from the wells by inverting the plate, and the wells have been washed twice with ice-cold PBS. After washing, the labelled cells have been resuspended in pre-heated medium at 5 × 105 cells/mL. 100 microliters of the labelled cells (50,000 cells) have been added to every nicely. The plate was incubated at 37 °C for 60 min to activate the receptor in order that it sure to the coated ligand. After the incubation, the plates have been inverted and pressed towards a paper towel, and 150 µL of ice-cold PBS was then slowly pipetted into the wells and the plate was gently shaken at a low velocity for 3 minutes. This was repeated one other two occasions, and 100 µL of PBS was added to the wells and the plate was analyzed with a Varioskan LUX multimode microplate reader (Thermo Fisher). The wavelengths used have been 480 nm for excitation and 502 nm for emission.
EV isolation
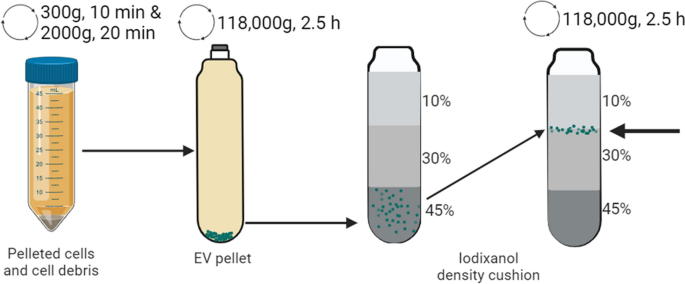
A schematic overview of the EV isolation course of is described in Fig. 1. Cells, cell particles, and huge apoptotic our bodies have been faraway from the cell tradition media through centrifugation at 300 × g for 10 min and 2000 × g for 20 min. EVs have been solely remoted from cultures rising in suspension in FBS-free media. The supernatant was ultracentrifuged for two.5 h at 118,000 × g at 4 °C (38,500 rpm, Kind 45 Ti mounted angle rotor, 181 as k-factor, Beckman Coulter). The pellet was resuspended in 0.5–1 mL PBS, and the EVs have been additional purified through a bottom-loaded iodixanol density cushion. The cushion was generated by mixing the pattern with 3 mL 60% iodixanol within the backside of the tube (remaining focus 45%) after which including 4 mL of 30% iodixanol and three mL of 10% iodixanol to this layer. The pattern was then ultracentrifuged at 100,000 × g for two h (28,000 rpm, SW 41 Ti swinging rotor, 265 as k-factor, Beckman Coulter). The purified EVs have been collected on the 10–30% iodixanol interphase.
Overview of the steps for the isolation and purification of the EVs. Cells, cell particles, and huge apoptotic our bodies have been eliminated by gradual centrifugation (300 × g and 2000 × g). The remaining EVs have been pelleted by ultracentrifugation (118,000 × g) and have been additional purified through a bottom-loaded density cushion with iodixanol (45%, 30%, and 10%)
EV characterization through transmission electron microscopy (TEM)
LFA-1 EVs have been negatively stained and visualized by TEM. Briefly, vesicles have been loaded onto glow-discharged 300-mesh copper grids (Electron Microscopy Sciences) for 30 s, then washed with water twice and additional stained with 2% uranyl formate for 1 min. Unfavorable-stained EVs have been analyzed by acquisition on a TALOS L 120 C transmission digital microscope (Thermo Fisher) at 120 kV with a BM-Ceta CMOS 4k*4k CCD digicam.
Western blot
The protein concentrations of cell lysates and EVs remoted from completely different clones was decided with a Pierce BCA Protein assay package (Thermo Scientific) following the advisable protocol. Samples for Western blot have been made by diluting 5 µg protein from the lysates or EVs to a quantity of 15 µL after which mixing it with 15 µL Pattern Buffer (Bio-Rad, 2× Laemmli pattern buffer), with or with out dithiothreitol (DTT), to a complete quantity of 30 µL. DTT was used when analyzing samples for CD11a however not for CD18. Samples have been added to Mini-PROTEAN TGX Stain-Free gels with seven wells (Bio-Rad) along with Precision Plus Protein Commonplace (Bio-Rad), utilizing 1× Tris/Glycine/SDS Buffer (Bio-Rad). Proteins have been transferred from the gel to a PVDF membrane (Bio-Rad) with Trans-Blot Turbo Mini-size Switch Stacks (Bio-Rad) and 1× trans-blot buffer (Bio-Rad Trans-Blot Turbo 5× Switch Buffer) utilizing a semi-dry switch chamber (Bio-Rad). The membrane was blocked with a blocking buffer (Bio-Rad EveryBlot Blocking Buffer) and incubated for 20 min. After this incubation, antibodies towards CD11a (ab186873, Abcam, 1:1000 dilution) or CD18 (MEM-48, Thermo Fisher Scientific, 1:1000 dilution) have been added and incubated in a single day at 4 °C. The membrane was then washed thrice with washing buffer (Bio-Rad 1× TBS with 0.1% Tween 20), incubated with the secondary antibody in blocking buffer for 60 min after which washed once more thrice with washing buffer. The membrane was then developed utilizing Supersignal West Femto Most sensitivity substrate (Thermo Fisher Scientific) on a Chemidoc Imaging System (Bio-Rad).
Nano-FCM of EVs
Floor molecule expression was assessed by nano-FCM utilizing a Movement NanoAnalyzer (NanoFCM Inc.) in accordance with the producer’s pointers. Previous to pattern loading, the Movement NanoAnalyzer underwent a calibration course of for alignment parameters as specified by the producer. This alignment concerned focus/high quality management (QC beads, NanoFCM Inc.), dimension beads (Silica nanospheres; 68–155 and 155–850 nm; NanoFCM Inc.), and a clean management. Per the producer’s directions, the measured occasions per minute have been maintained beneath 12,000 occasions. To realize this, samples have been initially assessed, and if the depend exceeded 2000 occasions within the first 15 s of measurement the pattern was appropriately diluted to keep up counts between 11,000 and 12,000 occasions.
For immunofluorescent staining, 1 µL of the prediluted antibodies at a 6-fold dilution was added to three µL of the diluted EV pattern, adopted by a 40-minute incubation at room temperature at nighttime. Subsequently, samples have been diluted 50 fold with PBS and promptly loaded onto the nano-FCM for information acquisition. The antibodies used for the staining step have been BD Pharmingen FITC mouse anti-human CD18 (Clone 6.7, BD Biosciences) and BD Pharmingen PE mouse anti-human CD11a (Clone HI111, BD Biosciences). The instrument parameters for the Movement NanoAnalyzer have been configured as follows: laser energy of 10 mW at 488 nm; laser energy of 20 mW at 638 nm; SS decay at 10%; sampling stress at 1.5 kPa; time to report at 1 min; and a 525/40 filter for FITC and a 488/30 filter for PE. The nano-FCM software program (NF occupation V1.0) was used to calculate the proportion of constructive sign, particle focus, and dimension distribution.
Proteomics—pattern preparation
The quantity of every pattern for the proteomic evaluation was normalized based mostly on the protein content material. Every pattern contained 40 µg protein (9 samples), and a reference pool was additionally constructed with contributions from all samples containing 40 µg in complete (4.44 µg/pattern). Sodium dodecyl sulfate (SDS) was added to all samples to a remaining focus of two%.
The samples and reference pool have been processed utilizing a modified filter-aided pattern preparation technique [18]. In brief, samples have been decreased with 100 mM DTT at 60 °C for 30 min, transferred to Microcon-30 kDa Centrifugal Filter Models (Merck), and washed a number of occasions with 8 M urea and as soon as with digestion buffer (DB; 50 mM TEAB and 0.5% sodium deoxycholate (SDC)) previous to alkylation with 10 mM methyl methanethiosulfonate in DB for 30 min at room temperature. Samples have been digested with trypsin (Pierce MS-grade trypsin, Thermo Fisher Scientific, 1:100 ratio) at 37 °C in a single day, and a further portion of trypsin was added and incubated for an additional 2 h. Peptides have been collected by centrifugation and labelled utilizing tandem mass tag (TMT) 11-plex isobaric mass tagging reagents (Thermo Fisher Scientific) in accordance with the producer’s directions. The samples have been mixed into one TMT-set and SDC was eliminated by acidification with 10% TFA. The TMT-set was additional purified utilizing a Excessive Protein and Peptide Restoration Detergent Removing Spin Column and Pierce peptide desalting spin columns (each from Thermo Fischer Scientific) in accordance with the producer’s directions previous to fundamental reversed-phase chromatography fractionation. Peptide separation was carried out utilizing a Dionex Final 3000 UPLC system (Thermo Fischer Scientific) and a reversed-phase XBridge BEH C18 column (3.5 μm, 3.0 × 150 mm, Waters Company) with a gradient from 3 to 100% acetonitrile in 10 mM ammonium formate at pH 10.00 over 23 min at a circulate of 400 µL/min. The 40 fractions have been concatenated into 18 fractions, dried, and reconstituted in 3% acetonitrile and 0.1% trifluoroacetic acid.
Proteomics—nanoLC-MS/MS evaluation and information base search
Every fraction was analyzed on an Orbitrap Lumos Tribrid mass spectrometer outfitted with the FAIMS Professional ion mobility system interfaced with an nLC 1200 liquid chromatography system (all from Thermo Fisher Scientific). Peptides have been trapped on an Acclaim Pepmap 100 C18 lure column (100 μm × 2 cm, particle dimension 5 μm, Thermo Fischer Scientific) and separated on an in-house-constructed analytical column (350 × 0.075 mm I.D.) full of 3 μm Reprosil-Pur C18-AQ particles (Dr. Maisch, Germany) utilizing a gradient from 3 to 80% acetonitrile in 0.2% formic acid over 85 min at a circulate of 300 nL/min. Precursor ion mass spectra have been acquired at 120,000 decision, scan vary 375–1375, and most injection time 50 ms. MS2 evaluation was carried out in a data-dependent mode, the place probably the most intense doubly or multiply charged precursors have been remoted within the quadrupole with a 0.7 m/z isolation window and dynamic exclusion inside 10 ppm for 45 s. The remoted precursors have been fragmented by collision-induced dissociation at 35% collision power with a most injection time of fifty ms for 3 s (‘prime velocity’ setting) and detected within the ion lure, adopted by multinotch (simultaneous) isolation of the highest 10 MS2 fragment ions inside the m/z vary 400–1400, and additional fragmentation (MS3) by higher-energy collision dissociation (HCD) at 65% collision power and detection within the Orbitrap at 50,000 decision, m/z vary 100–500, and a most injection time of 105 ms.
The information recordsdata for every set have been merged for identification and relative quantification utilizing Proteome Discoverer model 2.4 (Thermo Fisher Scientific). The search was towards Homo sapiens (Swissprot Database, April 2023, 20,422 entries) utilizing Sequest as a search engine with precursor mass tolerance of 5 ppm and fragment mass tolerance of 0.6 Da and Sequest HTXCorr set to 2. Tryptic peptides have been accepted with one missed cleavage, variable modifications of methionine oxidation, and glued cysteine alkylation, and TMT-label modifications of the N-terminus and lysine have been chosen. Percolator was used for PSM validation with the strict FDR threshold of 1%. TMT reporter ions have been recognized with 3 mmu mass tolerance within the MS3 HCD spectra, and the TMT reporter abundance values for every pattern have been normalized to the entire peptide quantity. Solely the quantitative outcomes for the distinctive peptide sequences with a minimal SPS match of 65% and a mean S/N above 10 have been thought of for the protein quantification. The reference samples have been used because the denominator and for the calculation of the ratios. The quantified proteins have been filtered at 1% FDR and grouped by sharing the identical sequences to attenuate redundancy.
Binding assay of LFA-1-expressing EVs to ICAM-1 cells
A binding assay was utilized to find out whether or not the membrane proteins CD11a and CD18 current on the EVs type a useful receptor. Wells in a 96-well plate (Thermo Scientific, Black microplate F96) have been coated with EVs from the LFA-1 clone (LFA-1 EVs) by including 10 µg/mL of the EVs in 100 µL PBS. Alternatively, wells have been coated with EVs remoted from the CD18 clone (CD18 EVs) or EVs remoted from WT HEK293F cells (WT EVs), and LFA-1 EVs that had been pre-incubated with 100 µg/mL anti LFA-1 neutralizing antibody (InVivoMAb, Clone TS-1/22.1.1.13). PBS alone was added to the wells as adverse controls. In these binding assays, labelled ICAM-1-expressing HMEC-1 cells have been added to the wells. The HMEC-1 cells have been handled with 15 ng/mL human tumor necrosis issue alpha (TNF-α; 210-TA-005, R&D programs) in a single day to extend ICAM-1 expression on the cells. The washing and blocking of the plates and labelling of the cells have been the identical because the earlier binding assay. After the labelled cells have been added to the wells, the plate was incubated for 30 min at 37 °C after which washed and analyzed as for the earlier binding assay.
Uptake experiment
100,000 HMEC-1 cells have been seeded in a 24-well plate (Falcon) (0.5 mL). After the cells had been allowed to stick (3–4 h), they have been handled with 15 ng/mL TNF-α (210-TA-005, R&D programs) and incubated in a single day. EVs have been stained with the fluorescent dye DiO by incubation for 30 min at 37 °C. The following day the cells have been washed with PBS after which incubated with an anti ICAM-1 neutralizing antibody (R&D Programs, clone BBIG-I1; utilizing concentrations 0.3–30 µg/mL) or media for 90 min at 37 °C. Following this incubation the cells have been once more washed and incubated with both 5 × 109/mL DiO-stained EVs or 5 × 109/mL revesiculated EVs loaded with the fluorescent peptide and incubated for 30 min within the incubator. The wells have been then washed twice with PBS and handled with 0.2 mL trypsin (Cytiva Hyclone) for five min at 37 °C. The trypsinated cells have been then transferred to fifteen ml Falcon tubes, diluted in 5 mL tradition medium, and pelleted through centrifugation at 300 × g for 7 min. Following the centrifugation, the supernatant was eliminated, and the cells have been resuspended in 500 µL fixation buffer (BD CellFIX) and incubated for 15 min at 4 °C. The mounted cells have been washed utilizing 1.5 mL FACS buffer (PBS with 1% FBS) adopted by centrifugation at 200 × g for 10 min. After the centrifugation the supernatant was eliminated, and the cells have been resuspended in 500 µL FACS buffer and analyzed through circulate cytometry by which 10,000 information factors have been collected. The information have been analyzed with FlowJo Software program (Tree Star Inc.).
Producing and loading EVs through the open-and-close process
To enhance the anti-inflammatory potential of the LFA-1 EVs, they have been loaded with the anti-Myd88 anti-inflammatory peptide through an open-and-close process to generate revesiculated EVs as beforehand described [15]. Anti-inflammatory peptides concentrating on Myd88 have been synthetized by JPT Peptide Applied sciences. Connected to the Cys facet chain was a fluorescent tag (Fluorescein), and the peptide sequence was Arg-Asp-Val-Leu-Professional-Gly-Tr-Cys-Val-Asn-Ser-cholesterol. This ldl cholesterol tag permits the peptide to affiliate with the EVs strongly.
The LFA-1 EVs (1011 EVs) have been handled with a excessive pH answer and incubated at room temperature for 30 min, after which the membrane sheets have been washed with PBS. After the washing, the membranes have been resuspended in 400 µL PBS with 50 µg of the peptide. The membranes have been incubated with the peptides at 37 °C for 30 min after which they have been sonicated for 30 min at room temperature. Peptide-loaded revesiculated EVs have been separated from non-loaded peptides by way of iodixanol-based ultracentrifugation, utilizing a gradient consisting of three mL of 60% iodixanol blended with the pattern, 4 mL of 30% iodixanol, and three mL of 10% iodixanol, at 100,000 × g for two h (28,000 rpm, SW 41 Ti swing rotor, 265 as k-factor, Beckman Coulter). The vesicles positioned within the interphase between the ten% and 30% iodixanol layers have been collected. A multimode microplate reader (Varioskan LUX, Thermo Fisher Scientific) was used to find out the peptide loading effectivity with excitation and emission wavelengths set at 495 nm and 520 nm, respectively. The quantification of peptides was decided based mostly on a calibration curve, enabling correct calculation of the peptide loading effectivity within the collected vesicles.
Evaluating the anti-inflammatory potential of LFA-1 EVMyd88
An assay was developed to find out how environment friendly LFA-1 EVMyd88 was at decreasing irritation in ICAM-1-expressing cells. HMEC-1 cells expressing ICAM-1 have been seeded out onto a 24-well plate (Falcon; 100,000 cells per nicely in 500 µL medium) and incubated in a single day. The tradition medium was changed with 500 µL recent medium together with TNF-α (3 ng/mL; 210-TA-005, R&D programs) to induce excessive ICAM-1 expression and inflammatory responses within the HMEC-1 cells. The pre-treatment with TNF-α was 3 h within the incubator after which the cells have been washed with PBS as soon as after which handled with both WT EVMyd88 or LFA-1 EVMyd88 (2 × 109/mL; 10,000 EVs per cell), with anti-Myd88 peptide because the constructive management (2 µg; corresponding quantity of peptide which was loaded in EVs) or PBS and never loaded LFA-1 EVs (2 × 109/mL; 10,000 EVs per cell) as adverse controls in 500 µL recent medium for 30 min. Following this therapy, the cells have been once more washed with PBS and 500 µL recent medium was added to the wells together with bacterial outer membrane vesicles (OMVs; 3 ng/mL) as a post-treatment to induce robust inflammatory responses. The OMVs have been remoted as beforehand described utilizing Escherichia coli pressure DH5α [15]. The post-treatment lasted 6 h, after which the supernatant was collected and the focus of the cytokine IL-8 was measured utilizing a DuoSet ELISA Growth package (R&D Programs, Minneapolis, MN).