Twin nanosystem engineering
Based mostly on the native mature sequence of miR-122 (miRBase ID: hsa-miR-122-5p), we designed a complementary 18-mer abasic PNA probe with an abasic place within the central area, represented as *GL*, containing a secondary amine and a glutamic acid-like aspect chain at its gamma place instantly reverse a guanine (underlined within the hsa-miR-122-5p sequence). The abasic PNA probe additionally accommodates an azide moiety through a spacer (two diethylene glycol items, miniPEG, proven as “OO“) to permit the clicking chemistry response. As well as, we’ve launched glutamic acid-like aspect chains on the gamma positions within the spine of sure PNA items (proven as Cglu and Tglu) to facilitate the templated DCL response between SMART-Nucleobases and the secondary amine of the abasic website [34]. We carried out DCL reactions in resolution utilizing the abasic PNA probe and totally different goal sequences equivalent to ssDNA-122, ssRNA-122 and hsa-miR-122-5p extracted and pre-amplified from tumour cell lysates (Fig. 1A), with all sequences listed in Desk 1. When added to the answer, the abasic PNA probe hybridises with the miR-122 goal, with the abasic place reverse a guanine. This guanine templates the incorporation of a biotinylated SMART-Cytosine (SMART-C-Biotin), forming a biotinylated PNA:miR-122 complicated (Fig. 1B). The chemical constructions of the abasic PNA probe and SMART-C-Biotin are proven intimately within the inset of Fig. 1B. Then, polystyrene nanoparticles, referred to as BCN-Cy5-NPs (12), are added to the answer. These nanoparticles have cyanine-5 (Cy5) fluorescent tags and are coated with a cyclooctyne moiety (BCN), which facilitates copper-free click on chemistry azide-labelled PNA probes, thereby capturing the biotinylated PNA:miR-122 complicated. Subsequent, silica nanoparticles, referred to as Strep-LINARDA (L3), are added to the answer. These nanoparticles are labelled with yellow-green emitting fluorophores just like cyanine-3 (Cy3) and coated with streptavidin to recognise the SMART-C-Biotin. Following the clicking chemistry response and streptavidin–biotin recognition, each nanoparticles, the BCN-Cy5-NPs (12) and Strep-LINARDA (L3), bind collectively via the PNA:miR-122 complicated, forming a twin nanosystem (Fig. 1C). Subsequently, the upper the quantity of miR-122 within the pattern, the upper the variety of biotinylated PNA:miR-122 complexes fashioned, and consequently, the upper the variety of twin nanosystems detected. When analysed with an ordinary movement cytometer, the twin nanosystem generates two co-localised fluorescent alerts, Cy5 + /Cy3 + , which served as markers for miR-122 detection with single base specificity (Fig. 1D). Additional particulars on the chemical construction of the essential PNA and all of the biotinylated SMART-Nucleobases may be present in Supporting Data 1. Furthermore, we carried out additional research on miR-122 sequences and extra targets, whose sequences are listed in Supporting Data 2.
Click on chemistry-based twin nanosystem workflow. A Hsa-miR-122-5p sequences are remoted from tumour cell strains and pre-amplified by PCR. B The addition of abasic PNA probes that hybridise to the miR-122 goal, and, along with the addition of the SMART-C-Biotin, the DCL response is initiated, ensuing within the formation of biotinylated PNA:miR-122 complexes. The inset exhibits the chemical constructions of the abasic PNA probe and SMART-C-Biotin. C Two forms of nanoparticles are then added: First, polystyrene nanoparticles, BCN-Cy5 NPs (12), labelled with Cy5 and coated with cyclooctyne (BCN) moieties. These nanoparticles permit copper-free click on chemistry to seize the PNA:miR-122 complicated. Second, silica nanoparticles, Strep-LINARDA (L3), that are Cy3-labelled and coated with streptavidin for recognition of the biotinylated SMART-Nucleobase. When each nanoparticles are certain by the PNA:miR-122 complicated, they kind the twin nanosystem. D Co-detection of Cy5 + /Cy3 + alerts utilizing an ordinary movement cytometer signifies detection of miR-122 with single base specificity
Synthesis, characterisation and validation of the twin nanosystem for miR-122 detection
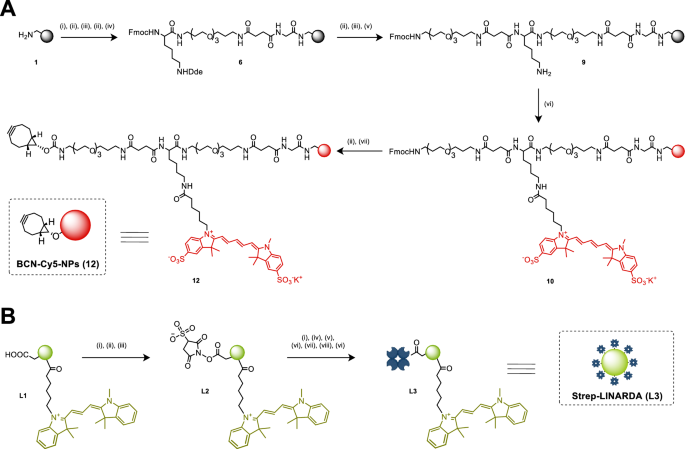
Within the synthesis of the elements of the twin nanosystem, the synthesis of polystyrene nanoparticles, BCN-Cy5-NPs (12), begins with the era of amino-functionalised PS-NPs (1), that are then bifunctionalised utilizing beforehand reported solid-phase synthesis strategies [24]. Monomers are conjugated to the amino PS-NPs (1) by activating the carboxyl group and forming amide bonds. These monomers have protecting teams, particularly Fmoc and Dde moieties, which permit stepwise and orthogonal synthesis. This permits bifunctionalisation with Cy5 fluorophores and BCN moieties, avoiding cross reactions (Fig. 2A). As soon as synthesised, BCN-Cy5 NPs (12) may be monitored by movement cytometry resulting from their Cy5 labelling. They’ll additionally conduct copper-free click on chemistry by reacting with azide-labelled PNA in resolution through the BCN moiety. The detailed synthesis for PS-NPs (1) is described in Supporting Data 3.1, whereas the total synthesis process for BCN-Cy5-NPs (12) is detailed in Supporting Data 3.2. The second sort of silica nanoparticles, Strep-LINARDA (L3), are carboxylic acid functionalised and labelled with Cy3-like fluorophores. They had been instantly conjugated to streptavidin by activating the carboxylic acid group with EDC and sulfo-NHS (Fig. 2B). The outcome was Strep-LINARDA (L3), which might recognise the biotinylated SMART-Nucleobases.
Nanoparticle synthesis. A Synthesis of BCN-Cy5 NPs (12) begins with amino-functionalised PS-NPs (1). Protected monomers Fmoc-Gly-OH, Fmoc-PEG-OH, Fmoc-Lys(Dde)-OH and Fmoc-PEG-OH are sequentially coupled. Subsequent, the Dde group is selectively eliminated to acquire to acquire PS-NPs (9) and the activated sulfo-Cy5-NHS ester is conjugated. The Fmoc group is then eliminated and the activated BCN succinimidyl ester is conjugated to acquire bifunctionalised BCN-Cy5-NPs (12). B Synthesis of Strep-LINARDA (L3) consist on the activation of the carboxylic teams utilizing EDC and sulfo-NHS to permit the conjugation of streptavidin to acquire the Strep-LINARDA (L3) nanoparticles
After the synthesis of each nanoparticles, their physicochemical characterisation was carried out. The dimensions and homogeneity of the nanoparticles and their polydispersity index (PDI) had been measured by Dynamic Mild Scattering (DLS), which confirmed an analogous nanoscale measurement and that each forms of nanoparticles had been monodispersed and homogeneous (Desk 2, Fig. 3A). So as to assess the change within the nanoparticle floor after the synthesis and its potential influence on floor stability, the zeta potential of the nanoparticles was additionally measured (Desk 2, Fig. 3A). A decrease zeta potential was noticed within the functionalised BCN-Cy5 NPs (12) and Strep-LINARDA (L3), in comparison with the PS-NPs (1), in all probability because of the presence of sulphonic teams, however these zeta potentials didn’t present a big change on the nanoparticle floor, indicating that the nanoparticles stay steady after conjugations. As well as, the effectivity of Cy5 conjugation to BCN-Cy5-NPs (12) and the conjugation of streptavidin to Strep-LINARDA (L3) had been evaluated, demonstrating profitable conjugation charges in each instances (Desk 2, Fig. 3B). Transmission electron microscopy (TEM) confirmed the morphology and measurement of each forms of nanoparticles (Fig. 3C). Further physicochemical characterisation was carried out to evaluate the scale, uniformity and zeta potential of the nanoparticles in all buffers used on this examine, together with in a serum matrix. Particulars of those outcomes are proven in Supporting Data 4.1.
Twin nanosystem characterisation and miR-122 detection by FACSVerse. A Measurement distribution (left) and zeta potential (proper) values for PS NPs (1), BCN-Cy5 NPs (12) and Strep-LINARDA (L3). B Calibration curves for Cy5 and streptavidin conjugation giving 96.45% and 99.7% conjugation charges respectively. C Measurement and morphology evaluation by TEM of Strep-LINARDA (L3) (left) and BCN-Cy5 NPs (12) (proper) (100 nm scale bar above, 500 nm scale bar beneath). D Dot plots exhibiting, on the left, particular person alerts generated by each nanoparticles. The highest-middle panel exhibits BCN-NPs (14) incubated with ssDNA-122-Cy5 with out the abasic PNA probe as a unfavorable management. Within the presence of the abasic PNA probe, within the decrease center panel, the sign turns into Cy5+, confirming the profitable click on chemistry response. The highest proper panel represents each nanoparticles within the presence of all elements however with out the ssDNA-122 goal as a unfavorable management, turning Cy5 + /Cy3 + within the presence of ssDNA-122 (in blue), highlighting miR-122 detection. E Specificity take a look at of the twin nanosystem evaluating the variety of occasions (n = 3) of various unfavorable management (NC) situations and within the presence of miR-122. A major improve within the Cy5 + /Cy3 + subset happens when the twin nanosystem is fashioned when ssDNA-122 is current. Error bars are expressed as ± SEM. Statistical significance was analysed utilizing one-way ANOVA, evaluating the imply of all NCs with the imply of miR-122. Significance is indicated as ****p < 0.0001
Then, we validated the power of the twin nanosystem to detect miR-122 utilizing a Fluorescence-Activated Cell Sorting cytometer, FACSVerse. When analysed on FACSVerse, the BCN-Cy5-NPs (12) confirmed a Cy5 + /Cy3- inhabitants, whereas the Strep-LINARDA (L3) confirmed a Cy5-/Cy3 + inhabitants (Fig. 3D, left). Then, we evaluated the efficacy and specificity of the clicking chemistry response. For this goal, we designed a 3rd sort of polystyrene nanoparticles that had been synthesised with out the Cy5 dye however conjugated to BCN, referred to as BCN-NPs (14) (see synthesis and construction in Supporting Data 3.3). We then used artificial ssDNA-122 labelled with the Cy5 fluorophore (ssDNA-122-Cy5) because the goal. Once we blended the BCN-NPs (14) with ssDNA-122-Cy5 within the absence of the abasic PNA probe that carries the azide group, no inhabitants exhibiting Cy5 fluorescence was detected (Fig. 3D, central black dot plot). Nonetheless, upon addition of the abasic PNA probe, the complete nanoparticle inhabitants confirmed Cy5 fluorescence (Fig. 3D, central crimson dot plot). This means profitable seize of the PNA:miR-122 complicated fashioned in resolution by hybridisation through a copper-free click on chemistry response. It additionally exhibits that polystyrene nanoparticles don’t adsorb oligonucleotides non-specifically. Subsequent, we evaluated the power of the twin nanosystem to detect miR-122. To do that, a DCL response was carried out in line with the process described in Sect. “DCL response and miR-122 detection with single-base specificity by FACS”. Once we evaluated the nanosystem within the absence of ssDNA-122 as a unfavorable management, a Cy5 + /Cy3- inhabitants was fashioned (Fig. 3D, proper gray dot plot). Remarkably, within the presence of ssDNA-122, there was a marked shift in the direction of a big double Cy5 + /Cy3 + inhabitants, indicating the profitable detection of ssDNA-122 (Fig. 3D, proper blue dot plot). Equally, once we analysed ssRNA-122, we noticed a big double Cy5 + /Cy3 + inhabitants (knowledge proven in Supporting Data 4.2). To check the specificity of the twin nanosystem, we then carried out an assay with 4 unfavorable controls in line with the experimental design proven in Desk 3. The result’s proven in Fig. 3E, displaying the variety of occasions (n = 3) recorded in FACSVerse for every situation and the statistical significance of the twin nanosystems within the detection of ssDNA-122 (statistical significance for ssRNA-122 is proven in Supporting Data 4.2). We carried out a further analysis of the twin nanosystem utilizing a distinct sort of silica nanoparticles, DiagNano™ (CD Bioparticles, UK), as a business different to Strep-LINARDA (L3). Our outcomes confirmed the profitable detection of miR-122 utilizing DiagNano™, demonstrating the flexibility of mixing the nanosystem with different nanoparticle varieties. These outcomes are offered in Supporting Data 4.3.
All outcomes demonstrated the profitable synthesis of each forms of nanoparticles, which exhibited homogeneity and stability, as confirmed by their physicochemical characterisation. Moreover, the outcomes indicated that the clicking chemistry response occurred with each excessive effectivity and memorable specificity, confirming the reliability of the synthesis course of. As well as, the outcomes confirmed that each forms of nanoparticles efficiently detected miR-122 with excessive specificity, which may be simply monitored by movement cytometry FACSVerse.
Detection of miR-122 in a serum matrix, calibration curve and analysis of single base specificity
Then, we proceeded to guage the power of the twin nanosystem to detect miR-122 in a organic matrix to problem the system. To do that, we carried out the next experiments in 10% of human serum. Below these situations, we established a calibration curve to find out the analytical efficiency and restrict of detection (LoD), and evaluated the specificity in detecting nucleotide variations. We established the calibration curve by performing triplicate dilutions of ssDNA-122 inside a variety masking from 50 pmol to 0.5 pmol in 10% of human serum in a remaining quantity of fifty µL. Then, we calculated the common variety of occasions throughout the double Cy5 + /Cy3 + inhabitants based mostly on a complete of 10,000 occasions recorded in FACSVerse. The ensuing common variety of occasions confirmed a sigmoidal regression curve (R2 = 0.9) (Fig. 4A, left) with a coefficient of variation starting from 0.9 to 9.22%, indicating low variability of the info. We then decided the LoD utilizing a 4-parameter logistic regression mannequin (4PL). First, the restrict of clean (LoB) was calculated utilizing components (1), after which this LoB worth was substituted into the variable “y” in components (2). The LoD was quantified as 2.329 pmol in 50 µL.
Calibration curve of miR-122 in serum matrix, SNV detection and MALDI-TOF evaluation. A Left, the calibration curve for the detection of ssDNA-122 in 10% serum confirmed a 4PL non-linear regression mannequin in FACSVerse. LoD = 2.329 pmol. R2 = 0.9. Proper, calibration curve for the detection of ssDNA-122-Cy5 and unlabelled BCN-NPs (14). LoD = 1.08 fmoles. R2 = 0.98. B FACSVerse evaluation of 4 SNVs representing 4 isomiR-122. Solely when the complementary biotinylated SMART-Nucleobase (proven as SC, SA, SG or ST) is used, the twin nanosystem generates a big Cy5 + /Cy3 + inhabitants. C MALDI-TOF evaluation was carried out on the 4 SNVs to guage the cross-reactivity between the SMART-Nucleobases. The spectra confirmed two principal peaks representing the unreacted abasic PNA probe (left) and the probe with the biotinylated SMART-Nucleobase integrated (proper). The distinction between these peaks indicated the molecular mass of the particular SMART-Nucleobase integrated, enabling identification of the goal nucleobase. Error bars are expressed as ± SEM. Calibration curves had been analysed utilizing a 4PL mannequin. Statistical significance was assessed by one-way ANOVA. Significance is indicated as *p < 0.05, **p < 0.01, ***p < 0.001
$$LoB=Clean; Common; (n=6)+3.3*S{D}_{Common}$$
(1)
$$y=Backside+ frac{Prime-Backside}{1+{left(frac{x}{IC50}proper)}^{HillSlope}}$$
(2)
We discovered that the sign from the clean corresponds, largely, to the sign generated by the non-specific interplay between BCN-Cy5-NPs (12) and Strep-LINARDA (L3), therefore impacting within the LoD. To research whether or not the twin nanosystem may obtain a decrease LoD within the absence of this non-specific sign, we analysed the detection of dilutions of ssDNA-122-Cy5 utilizing the unlabelled BCN-NPs (14). Thus, optimistic Cy5 alerts throughout the double optimistic Cy5 + /Cy3 + inhabitants replicate solely the quantity of ssDNA-122-Cy5 captured by click on chemistry, excluding any non-specific sign. When analysing a 4PL, we discovered that the LoD was 1.08 femtomoles in 50 µL (Fig. 4A, proper), which is greater than 2,000 orders of magnitude decrease, demonstrating the potential room for enchancment of this twin nanosystem.
At low concentrations, the nanosystem displays non-linearity, because of the intrinsic nature of the processes concerned, as each the hybridisation response and fluorescence emission don’t comply with linear kinetics. For hybridisation, effectivity will increase because the goal focus approaches a threshold the place ample binding occasions generate a detectable sign. Equally, fluorescence emission requires a signal-to-noise ratio above background to confidently establish the goal sign. Collectively, these elements decide the detection threshold. Subsequent, we investigated the power to discriminate SNVs. For this finish, the abasic PNA probe was hybridised in triplicate to 4 ssDNA-122 strands with sequences differing by just one nucleotide (A, C, G or T) within the place reverse to the abasic website (Desk 1), thus representing 4 isomiR-122 variants. For every sequence, 4 particular person DCL reactions had been carried out in 10% serum utilizing one of many biotinylated SMART-Nucleobases (SMART-C-biotin, SMART-A-biotin, SMART-G-biotin, or SMART-T-biotin). Then, we analysed the outcomes obtained in FACSVerse and confirmed that solely the particular SMART-Nucleobase resulted in a statistically vital double Cy5 + /Cy3 + inhabitants sign (Fig. 4B). These outcomes demonstrated the power of the twin nanosystem to detect the particular SNV at a focused nucleobase place throughout the miR-122 sequence. To additional validate the twin nanosystem and broader applicability, we additionally used two further abasic PNA probes to analysed two nucleotide positions in miR-21, one other miRNA identified for its position in tumour growth [35]. These knowledge are proven in Supporting Data 4.4. With these outcomes, we additional verify the potential for utilizing the twin nanosystem to check further targets, in addition to the power to check different nucleobase positions.
To additional analyse the specificity of the biotinylated SMART-Nucleobases and whether or not there was any cross-reactivity between them, we carried out MALDI-TOF evaluation. For this, 4 particular person DCL reactions had been carried out, every utilizing one of many 4 ssDNA-122 sequences differing by one nucleotide (Desk 1). Every ssDNA-122 was blended in an answer containing the abasic PNA probe and a combination with all 4 biotinylated SMART-Nucleobases. The product was then analysed by MALDI-TOF, which revealed two principal peaks within the spectra: one peak similar to the unreacted abasic PNA probe and a second peak similar to the abasic PNA probe plus the biotinylated SMART-Nucleobase. The distinction between these peaks corresponds to the molecular weight of the integrated SMART-Nucleobase. The outcomes confirmed that solely the complementary SMART-Nucleobase was efficiently integrated into the abasic place, with no cross-reactivity noticed (Fig. 4C). We additionally carried out additional MALDI-TOF analyses to confirm the detection of SMART-T-Biotin, resulting from its molecular weight being near that of SMART-C-Biotin, as detailed in Supporting Data 5.1. As well as, we additionally examined cross-reactivity utilizing 4 templates of ssRNA-122 that differ in only one nucleotide (5′UGGAGUGU-G/A/C/U-ACAAUGGUGUUUG3′), and obtained related outcomes when the template was ssRNA-122. These outcomes are offered in Supporting Data 5.2. All outcomes offered on this part verify that the twin nanosystem utilizing FACSVerse detects miR-122 in a organic matrix with specificity and robustness, and demonstrates its potential to discriminate with excessive accuracy between 4 miR-122 sequences differing by just one nucleotide.
Evaluation of hsa-miR-122-5p expression in tumour cell strains
Subsequent, we investigated the expression of miR-122 in several tumour cell strains utilizing the twin nanosystem and in contrast the outcomes with miR-122 expression utilizing a TaqMan assay, a well-established methodology for finding out miRNA expression in cell strains, commercially out there from Thermo Scientific (USA). On this assay, we mixed the twin nanosystem with qPCR, the place the RNA is reverse transcribed into complementary DNA (cDNA) after which amplified, producing a DNA template for the DCL response. This strategy has the benefit of elevated stability and longer pattern storage occasions. Subsequently, within the earlier Sect. “Detection of miR-122 in a serum matrix, calibration curve and analysis of single base specificity”, we primarily characterised the twin nanosystem utilizing ssDNA-122 as a substitute of ssRNA-122. We first in contrast the 2 strategies by producing a calibration curve utilizing the total sequence of ssRNA-122 (5′ UGGAGUGUGACAAUGGUGUUUG 3′) because the beginning goal. We carried out a collection of dilutions in triplicate, utilizing quantities of ssRNA-122 starting from 1 ng to 10–8 ng, corresponding to eight × 1010 to eight × 102 copies of ssRNA-122. The RNA was then reverse transcribed into cDNA and subjected to RT-qPCR utilizing TaqMan probes. The results of plotting these portions in opposition to the cycle threshold (Ct) demonstrated a linear regression (R2 = 0.995) with coefficients of variation starting from 0.25 to 7% (Fig. 5A, left). The identical PCR merchandise had been then subjected to the twin nanosystem and analysed by FACSVerse. On this case, the results of plotting these portions in opposition to the variety of Cy5 + /Cy3 + occasions yielded a sigmoidal regression curve (R2 = 0.968) with coefficients of variation starting from 1.25 to 25% (Fig. 5A, proper). The LoD was then decided for each strategies. For the TaqMan assay, the LoD was calculated as 3.29 occasions the usual deviation of the unfavorable management divided by the slope of the calibration curve. The LoD was quantified as 5 copies in a remaining response quantity of 20 µL, corresponding to eight.30 × 10–24 mol (8.3 ymoles) of miR-122. The LoD of the twin nanosystem was then calculated utilizing the 4PL mannequin described in the earlier part. The LoD was quantified as 750 copies in a remaining quantity of fifty µL, similar to 1.3 × 10–21 mol (1.3 zmoles) of miR-122.
Evaluation of miR-122 expression in tumour cell strains utilizing TaqMan probes and the twin nanosystem. A Calibration curves of each strategies in RT-qPCR merchandise of ssRNA-122. On the left, the TaqMan probes present a linear regression curve. On the correct, the twin nanosystem exhibits a sigmoidal regression curve. B Evaluation of hsa-miR-122 expression in tumour cell strains. On the left, the TaqMan assay confirmed excessive hsa-miR-122 expression in HuH-7. Equally, on the correct, the twin nanosystem confirmed excessive hsa-miR-122 expression in HuH-7, adopted by a decrease expression in HepG2 and little to no expression in A549 or SKBR3. Error bars are expressed as ± SEM. The calibration curve of the twin nanosystem was analysed utilizing a 4PL mannequin. Expression evaluation utilizing TaqMan probes was carried out by calculating a Z-score evaluating the expression of every particular cell line to the common expression of miR-122 throughout the set. Statistical evaluation of miR-122 expression was carried out utilizing one-way ANOVA
Then, we analysed hsa-miR-122 expression in tumour cell strains. Since hsa-miR-122 is particular for liver tissue, we selected two human hepatocellular carcinoma cell strains: HuH-7, which has been reported within the literature to have excessive ranges of miR-122 expression, and HepG2, which has been reported to have a decrease miR-122 expression [36, 37]. We additionally chosen two further most cancers cell strains of medical curiosity for finding out miR-122: A549, derived from lung most cancers [38], and SKBR3, derived from breast most cancers, which in line with the literature has little or no expression of miR-122 [38, 39]. We cultured the cell strains to confluence and extracted complete miRNA from a million cell pellets in triplicate. For reverse transcription, 10 ng of RNA from every pattern was transformed to cDNA utilizing the TaqMan MicroRNA Reverse Transcription Package, which makes use of stem-loop primers particularly designed for microRNA cDNA synthesis [40]. Subsequently, 1.33 µL of the ensuing cDNA was subjected to qPCR in triplicate utilizing TaqMan Quick Superior Grasp Combine and particular TaqMan probes for hsa-miR-122 (ID 002245) and RNU44 (ID 001094). RNU44 was chosen as a normalisation management resulting from its steady expression in several human cell strains, guaranteeing dependable normalisation of microRNA expression knowledge [41]. Then, we analysed the identical amplicons utilizing the twin nanosystem through FACSVerse.
The outcomes confirmed that utilizing each strategies, we discovered excessive miR-122 expression within the HuH-7 cell line, adopted by decrease expression in HepG2, decreased expression in SKBR-3 and little to no expression in A549 (Fig. 5B). Extrapolating from the calibration curve, within the TaqMan assay, we discovered an expression of 4.51 × 106 copies of miR-122 in HuH-7 and three.47 × 103 copies in HepG2. In the meantime, with the twin nanosystem, we discovered 3.69 × 106 copies in HuH-7 and three.22 × 104 copies in HepG2. These outcomes show the analytical functionality of the twin nanosystem to analyse hsa-miR-122 expression in tumour cell strains, figuring out as much as 750 copies (1.3 zmol) in 50 µL (26 attomolar) quantity when mixed with PCR. Though the calibration curve confirmed increased variability at high values in comparison with the TaqMan assay, we attribute this to the truth that the movement cytometer might interpret a cluster of nanoparticles as a single occasion. Nonetheless, this didn’t have an effect on the accuracy of the miR-122 expression evaluation of the cell strains. Remarkably, the twin nanosystem outcomes correlated extremely with the TaqMan assay. This highlights the effectiveness of the twin nanosystem in performing miR-122 expression evaluation, whereas sustaining methodological robustness, compatibility with normal movement cytometers as a readily accessible platform, and the power to analyse SNVs and truncated miRNA sequences.