Rational screening of SHV-specific epitope
Since its inception, epitope imprinting has been adopted by several types of purposes. Nonetheless, to fulfil the imprinted materials specificity to focus on protein or protein household (based mostly on epitope recognition), query concerning which form of epitope to make use of as template have to be first answered. At the moment, two generally used approaches have been utilized to establish protein’s epitope. The best one is direct choice of the terminal sequence because the epitope [32], and the ready materials can straight adsorb the goal protein as a result of the chosen epitope is linear and uncovered. Nonetheless, the terminal sequence tends to mutate, and as soon as this occurs, the specificity of the imprinted materials will probably be misplaced. To deal with this deficiency, one other method emerged lately, through which a non-terminal peptide sequence is chosen because the epitope [33, 34], which defines that the sequence is uncovered and has a linear or easy loop construction relatively than an α-helix or β-sheet. This method extremely depends on the prediction and evaluation of the protein and peptide construction, and subsequently is sophisticated. Utilizing this method, though the ready materials can straight adsorb protein, the specificity for the goal protein or protein household could also be not passable.
As early as 2012, Bossi et al. proposed an method of protein fingerprint evaluation for identification of protein’s epitope [27]. This method depends on a easy bioinformatic evaluation, and it was confirmed to be efficient by later research [28]. Usually, three fundamental steps are concerned: (i) in silico cleavage of the goal protein by a given protease (e.g., trypsin, thermolysin, or enterokinase); (ii) choice of peptide fragments of ample size to execute a attribute evaluation (together with stability, hydrophilicity, and liposolubility); (iii) alignment of the filtered peptides towards protein databases to seek for sequence similarity utilizing the BLAST instrument. In the end, a novel and conserved peptide is recognized because the goal protein’s epitope, and the ready imprinted materials presents good specificity to the goal protein and parental household. For this method, it’s clear that the outlined epitope is extremely correlated with the given protease. If the epitope is uncovered outdoors of the protein, it may be straight adsorbed by the fabric; whether it is hidden contained in the protein, a protein pretreatment have to be carried out to reveal the epitope.
In contrast with the opposite two epitope-defined approaches, the method of protein fingerprint evaluation is handy, simple to know, and favorable to standardize the process of screening particular epitopes for goal proteins. On this examine, through the use of this method, a novel 10-mer peptide was recognized because the SHV-specific epitope, which was in vitro synthesized and used because the template for MEI-GP preparation. At first, we tried to straight adsorb SHV by MEI-GP, however sadly, no protein was adsorbed on materials. That is attributed to its non-linear native conformation and partially-hidden spatial place of the 10-mer peptide in authentic SHV molecule (Fig. 1C, particulars described within the following part). Subsequently, earlier than materials adsorption, a easy protein denaturation therapy was exerted on SHV molecules to get rid of the sorption obstruction derived from the epitope authentic construction. After that, the MEI-GP offered anticipated particular adsorption for SHV.
Preparation and recognition mechanism of MEI-GP
Synthesis and hydrophilic modification of Fe3O4 particle
A protein hydrodynamic diameter in aqueous resolution is normally roughly 5–10 nm [35]. Subsequently, as a substrate of the MEI-GP, the Fe3O4 particle ought to have a submicron measurement to offer a adequate adhesion floor for protein adsorption. Subsequently, a controllable solvothermal technique was used for Fe3O4 synthesis (Fig. 2), through which the particle measurement and the magnetic saturation worth had been tuned by the Fe3+ focus and the PEG dosage, respectively [31]. To enhance the water solubility of Fe3O4, a PAA modification was utilized after Fe3O4 synthesis (Fig. 2), through which the carboxy (–COOH) on PAA chain strongly coordinated to iron cations to type a sturdy coating, whereas uncoordinated –COOH prolonged into the answer, conferring Fe3O4 a excessive diploma of monodispersity within the water [36]. Nonetheless, the PAA modification is a double-edged sword, as a better PAA dosage or a bigger molecular weight prompted Fe3O4 to agglomerate. On this examine, 0.72 g of PAA 3000 (equimolecular with FeCl3) maximized Fe3O4 dispersion in aqueous resolution (Fig. 3A).
SiO2 coating on floor of Fe3O4–COOH particle
For SiO2 coating, three essential parameters had been individually optimized, together with TEOS dosage, response time, and NH3⋅H2O quantity. We discovered that the upper TEOS dosage and longer response time didn’t noticeably enhance the thickness of SiO2 membrane; nevertheless, they prompted Fe3O4@SiO2 agglomeration. Solely the NH3⋅H2O quantity was the important thing issue affecting SiO2 thickness. On this examine, utilizing 1 mL of NH3⋅H2O (25–28 wt%) catalyzation and including 3 mmol TEOS to the response in six equal parts with an interval time of 1 h, an SiO2 membrane with a thickness of three–5 nm was coated on floor of Fe3O4–COOH particle (Fig. 3B), and monodispersed Fe3O4@SiO2 was obtained. After SiO2 coating, a considerable amount of silicon hydroxy (–Si–OH) coated all the floor of Fe3O4@SiO2 (Fig. 2), guiding the particle participation in subsequent imprinting polymerization.
Imprinting polymerization occurring on floor of Fe3O4@SiO2
A easy and catalyst-free sol–gel polymerization was used for MEI-GP preparation, which was carried out at 30 °C, utilizing three silane coupling brokers, APTES, UPTES, and BHEAPTES, as useful monomers, and TEOS as cross-linker. After 3 h of incubation, the template epitope was captured by useful monomers; particularly, surrounding the template, an space wealthy in useful monomers was fashioned (Fig. 2). Derived from the hydrolysis of amino group (–NH2) on APTES and UPTES, this space contained loads of OHˉ, and was adequate to catalyze the hydrolysis of silicon ethoxy (–Si–OC2H5), adopted by the condensation response of –Si–OH. We discovered that when decreasing the dosage of useful monomers, the low-produced OHˉ couldn’t change –OC2H5, and the –Si–OH was not in a position to be produced; whereas high-produced OHˉ originating from high-dosage useful monomers inhibited the hydrolysis of ⋅OC2H5, and the cumulated ⋅OC2H5 restrained the manufacturing of –Si–OH. Thus, the affect of the useful monomer dosage on the sol–gel polymerization adopted a parabolic sample.
For imprinting optimization, we mounted dosages of useful monomers and cross-linker, and template epitope was adjusted inside a set molar ratio vary of template/APTES/UPTES/BHEAPTES/TEOS = (0.25–3):(5⋅8):(9⋅4):(3⋅4):(20⋅4), and several other epitope-imprinted polymers had been ready. We discovered that the sorption quantities of those supplies tapered off because the template dosage elevated (Fig. S1). When a low-dosage template was used for epitope imprinting, the fabric primarily confirmed non-specific adsorption, and when a high-dosage template was used, the polymer overcrosslinked and prompted many imprinted cavities coupled with template molecules to be trapped inside polymer community construction. Compared, a ratio of 1:(5⋅8):(9⋅4):(3⋅4):(20⋅4) was used because the optimum situation for MEI-GP preparation. Below this situation, the sorption quantity of the ready materials was steady, with solely small fluctuations occurring when template dosage was adjusted up and down. Herein, the molar ratio of 1:(x⋅8):(y⋅4):(z⋅4):((x + y + z)⋅4) can function a reference ratio for different epitope imprinting (x, y, and z, are the variety of –COOH, –CONH–, and –NH2 teams in template molecule that individually work together with –NH2, –CONH–, and –OH in useful monomer), which researchers can use to search out the optimum circumstances for his or her imprinted polymers.
With out areas wealthy in OHˉ (because of the absence of template epitope), the MNI-GP preparation required the assistance of NH3⋅H2O for catalysis. Just like the formation of MEI-GP, the affect of the NH3⋅H2O quantity on non-imprinting polymerization additionally adopted a parabolic sample.
Attributable to its participation in sol–gel polymerization of the –Si–OH on Fe3O4@SiO2 floor, the obtained MEI-GP and MNI-GP each had a mononuclear core–shell construction, with an imprinted and non-imprinted movie thickness of two–4 nm (Fig. 3C and D).
Recognition mechanism of MEI-GP for template epitope and SHV
For template epitope recognition by MEI-GP, two non-covalent interactions had been concerned, together with electrostatic attraction and hydrogen bonding (Fig. 2), which occurred in imprinted cavities. The imprinted cavity utterly matched the template when it comes to form, measurement, and positioning of chemical teams, which is the muse of epitope-specific recognition. The template VDAGDEQLER has good water solubility, indicating that it may be simply dissolved in polar solvents (e.g., 1,2-propanediol) however difficultly in low-polar liquids (e.g., ethanol). As a powerful polar solvent, water can simply disturb hydrogen bonding between chemical teams. Subsequently, on this examine, a combined solvent of ethanol/1,2-propanediol (1:1, v/v) was used because the porogen for MEI-GP preparation, and the ethanol/water (1:1, v/v) was used for template adsorption, in order that the non-covalent interactions may very well be expressed effectively.
Primarily based on epitope-specific recognition, SHV was selectively adsorbed by MEI-GP. Nonetheless, earlier than adsorption, two components had been considered. One is that the in vitro synthesized 10-mer epitope peptide tends to take care of a linear construction, which is completely different from its native conformation (α helix and Ω loop, Fig. 1C) in authentic SHV molecule. The opposite is that the epitope peptide partially hides within the globular construction (Fig. 1C) of the unique SHV, which might impede materials adsorption. Subsequently, the spherical SHV wanted to be denatured to a soluble linear construction earlier than adsorption, to make sure the epitope was utterly uncovered. For this, 2% (w/v) SDS was added to the protein resolution, which was heated at 100 °C for 10 min, and the denatured SHV was enwrapped by loads of anions, and subsequently grew to become soluble in aqueous resolution (Fig. 2). As well as, as a result of ethanol has a powerful capability to precipitate proteins, a medium of 1,2-propanediol/water (1:1, v/v) was used for SHV adsorption on this examine.
Characterizations
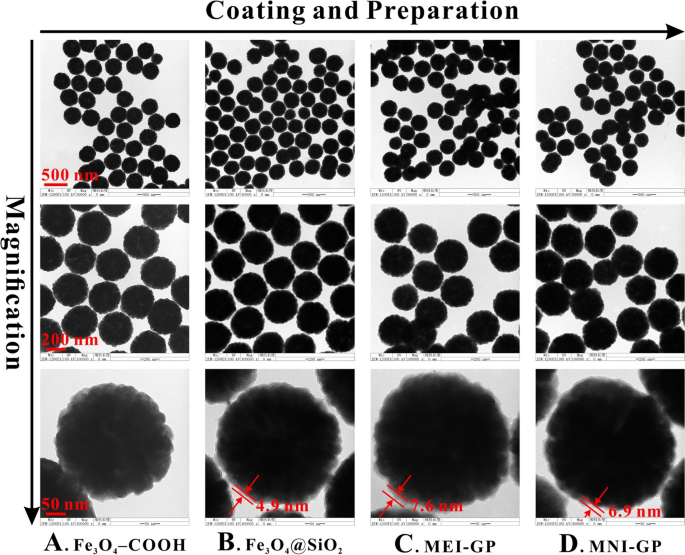
As proven in Fig. 3, the 4 supplies all had an almost spherical form, a slim measurement distribution (340 ± 20 nm), and a good monodispersed state in aqueous resolution. The Fe3O4 microsphere was composed of quite a few small nanoparticles, which endowed Fe3O4 with a polycrystalline nature [31]. The opposite three supplies all exhibited a mononuclear core–shell construction, with the shell thickness lower than 10 nm.
As will be seen in Fig. 4A, the C=O stretching vibration peak at 1715 cm−1 indicated the existence of –COOH on Fe3O4 floor. The absorption peak at 1635 cm−1 within the 4 spectra was ascribed to the deformation vibration of –OH of the 4 supplies. The vibration peak at 1564 cm−1 demonstrated the presence of –NH2 in silica membrane of MEI-GP and MNI-GP. Apart from Fe3O4–COOH, the opposite three supplies all had an Si–O–Si stretching vibration peak at 1095 cm−1. All the supplies had an apparent Fe–O peak at 580 cm−1, which was derived from the Fe3O4 substrate.
As proven in Fig. 4B, the binding energies of Fe 2p, O 1s, N 1s, C 1s, Si 2s, and Si 2p had been all clearly revealed. For Fe3O4–COOH, the O 1s and C 1s peaks at 531.3 and 288.3 eV had been simply discovered. For the opposite three supplies, the Si 2s and Si 2p peaks at 154.4 and 103.3 eV had been attributed to SiO2 coated on Fe3O4 floor. In contrast with Fe3O4@SiO2, each the visibly elevated C 1s peak (285.1 eV) and the looks of the N 1s peak (399.7 eV) in MEI-GP and MNI-GP spectra confirmed the profitable modification of useful teams of the 2 supplies.
As proven in Fig. 4C, the 4 supplies had the identical XRD sample, and the diffraction peaks at 30.1°, 35.5°, 37.1°, 43.1°, 53.5°, 57.1°, and 62.6° referred to the [220], [311], [222], [400], [422], [511], and [440] crystal planes, respectively, of the cubic inverse spinel Fe3O4. The place and relative depth of those diffraction peaks had been in good settlement with these from ICDD Ref. Code 01-075-0033 for Fe3O4, indicating that the crystallinity of Fe3O4 was steady throughout its useful modifications.
As proven in Fig. 4D, the magnetization curves of the 4 supplies had been utterly closed and symmetrical concerning the origin, with retentivity < 1 emu/g and coercivity < 20 Oe, demonstrating their typical superparamagnetic properties. The obtained MEI-GP had a excessive saturated magnetization of 52.9 emu/g, indicating that it may very well be conveniently separated from matrices by an exterior magnetic area.
Sorption behaviors of MEI-GP for template epitope and recombinant SHV
Binding isotherms
The binding isotherms of MEI-GP for template epitope and recombinant SHV are proven in Fig. 5A and B, respectively. Each used MNI-GP sorption for comparability. The sorption information had been fitted by three generally used fashions (Fig. 5C–E), the Langmuir, Freundlich, and Sips equations [36], that are expressed as follows, respectively:
$${textual content{Q}}_{{textual content{e}}} { = }frac{{{textual content{Q}}_{{textual content{m}}} {textual content{bC}}_{{textual content{e}}} }}{{{textual content{1 + bC}}_{{textual content{e}}} }}$$
(1)
$${textual content{Q}}_{{textual content{e}}} {textual content{ = Q}}_{{textual content{m}}} {textual content{C}}_{{textual content{e}}}^{{frac{{1}}{{textual content{n}}}}}$$
(2)
$${textual content{Q}}_{{textual content{e}}} { = }frac{{{textual content{Q}}_{{textual content{m}}} left( {{textual content{kC}}_{{textual content{e}}} } proper)^{{upgamma }} }}{{{1 + }left( {{textual content{kC}}_{{textual content{e}}} } proper)^{{upgamma }} }}$$
(3)
the place Qe is the equilibrium sorption quantity; Ce is the equilibrium focus of analytes in sorption medium; and Qm represents the sorption capability of the fabric for template epitope and recombinant SHV. The three constants, b, n, and ok, are associated to sorption vitality that depicts materials affinity to focus on analytes. The fixed γ is a dimensionless exponent that characterizes the sorption heterogeneity of the ready materials.
A, B Binding isotherms of MEI-GP and MNI-GP for template epitope (SHV-specific epitope) and recombinant SHV, utilizing TEM-specific epitope and recombinant TEM-1 as reference molecules. Earlier than adsorption, the spherical protein molecules (SHV and TEM-1) had been denatured to a linear construction. C–E Fitted curves of fabric sorption for goal analytes. The perfect matched mannequin is marked with a examine (seen within the dash-framed field). F, G Kinetics curves of MEI-GP for template epitope and recombinant SHV-1, which had been each effectively fitted by the pseudo-second-order mannequin (insert photos). The preliminary focus of the 2 analytes was 0.5 mg/mL
The Langmuir mannequin assumes that sorption happens on a skinny monolayer and all the binding websites are equal, which is commonly relevant for modeling the sorption occurring on homogeneous floor websites. The Freundlich mannequin assumes that sorption happens on thick multilayers and completely different websites with a number of sorption energies are concerned, which is extensively used for modeling the sorption occurring on heterogeneous binding websites. The Sips mannequin assumes that sorption first happens on monolayer binding websites, after which a multilayer sorption based mostly on molecular interplay takes place on materials floor.
As will be seen in Fig. 5C and Desk 2, materials sorption for template epitope was effectively fitted by Langmuir mannequin (R2 = 0.9838–0.9844), demonstrating that the binding websites on MEI-GP and MNI-GP surfaces had been each homogeneous and monolayer-distributed. In contrast with MNI-GP, the precise imprinted cavities endowed MEI-GP with a lot increased sorption capability (Qm = 37.71 mg/g) and a stronger sorption affinity (b = 16.21) for template epitope, and a good imprinting issue (Qm–MEI-GP/Qm–MNI-GP = 3.25) was additionally calculated.
As proven in Fig. 5D and E and Desk 2, Sip mannequin was best suited for becoming SHV (denatured) adsorption (R2 = 0.9818–0.9919), indicating that after the linear SHV was captured by the 2 supplies (monolayer adsorption), a van der Waals interplay happened between the adsorbed and free SHV molecules (multilayer overlaying). As a result of particular adsorption, MEI-GP exhibited a greater sorption impact (Qm = 27.41–28.66 mg/g, ok = 30.64–36.61) than MNI-GP for the 2 linear SHV.
As proven in Desk 2, a stronger affinity was noticed with SHV binding on MEI-GP, whereas increased sorption quantity occurred when template epitope was adsorbed by MEI-GP. This phenomenon was ascribed to the next causes (which had been partially revealed by different examine [37]): (i) After SHV captured by MEI-GP based mostly on epitope-specific recognition, the hydrogen bonding between the non-epitope a part of SHV and the –Si–OH on MEI-GP contributed SHV a stronger affinity. (ii) The big footprint of the captured SHV (286-mer) blocked adjoining binding websites (imprinted by a small epitope peptide (10-mer)), and subsequently fewer SHV may very well be sure on MEI-GP.
Selectivity
As proven in Fig. 5A, sorption quantity of the template epitope VDAGDEQLER sure on MEI-GP was a lot increased than that of the reference peptide VDAGQEQLGR (TEM-specific epitope), indicating that solely two amino acid substitutions considerably decreased peptide binding, which confirmed the precise recognition mechanism (form and measurement matching and useful group interactions) of MEI-GP.
As will be seen in Fig. 5B, attributable to epitope-specific recognition, MEI-GP exhibited higher selectivity for SHV than for the reference protein (recombinant TEM-1), indicating its capability to purify goal proteins from advanced matrices. As a result of MNI-GP had no imprinted cavities, its sorption for goal analytes was similar to that for reference molecules.
Sorption kinetics
As proven in Fig. 5F, the sorption fee of MEI-GP for template epitope was very quick and equilibrium was achieved in solely 2 min, because of the floor adsorption and utterly matched binding websites to the template. In distinction, as will be seen in Fig. 5G, the sorption fee for SHV-1 was slower and required almost 30 min to realize an preliminary equilibrium. This was attributed to a time-consuming epitope recognition course of and a size-exclusion impact between macromolecules (286-mer proteins) when competitively sure to micromolecule (10-mer peptide)-imprinted websites. As additionally proven in Fig. 5G, the sorption of SHV-1 elevated way more slowly after 30 min, indicating that formation of the van der Waals interactions (occurring after monolayer adsorption) between the adsorbed and free SHV molecules was a tardy course of.
The 2 kinetics curves had been fitted by a pseudo-second-order mannequin [36] (insert photos in Fig. 5F and G), which is expressed as:
$$frac{{ {textual content{t}}}}{{ textual content{q}_{textual content{t}} }} = frac{1}{{ nu_{o} }} + frac{{ {textual content{t}}}}{{ textual content{q}_{textual content{e}} }} = frac{1}{{textual content{ok}_{2}{ textual content{q}_{textual content{e}}^{2} } }} + frac{{textual content{t}}}{{textual content{q}_{textual content{e}} }}$$
(4)
the place qt and qe are sorption quantities of MEI-GP for goal analytes at time t and equilibrium, respectively; νo represents the preliminary sorption fee; and ok2 is a sorption fee fixed. This mannequin assumes that sorption fee is managed by chemisorption and the sorption capability depends on the variety of energetic binding websites on materials.
As proven within the insert photos in Fig. 5F and G and Desk 3, the pseudo-second-order mannequin fitted the 2 curves very effectively (R2 = 0.9992–0.9999), confirming that MEI-GP sorption for the 2 analytes was dominated by chemisorption: (i) For template epitope, non-covalent interactions with the imprinted cavities had been concerned. (ii) For SHV, each epitope-specific recognition and the next van der Waals forces contributed materials sorption.
Particular detection of SHV in micro organism
Below optimum circumstances (Fig. S2 and S3), SHV was selectively extracted from micro organism by MEI-GP after which submitted to MALDI-TOF MS for particular willpower (Fig. 6). As proven in Fig. 7 and Fig. S4 and S5, after SHV extracted by MEI-GP, matrix interferences had been successfully eradicated and the spectrum baseline grew to become very clean (in contrast with direct detection of bacterial crackate resolution), enabling SHV sign to be drastically enhanced and clearly offered in mass spectra. Though matrices had been additionally eliminated by MNI-GP, the SHV peak couldn’t be discovered within the spectra (i.e., MNI-GP couldn’t adsorb SHV from bacterial crackate resolution), demonstrating that epitope-specific recognition by MEI-GP was the important thing issue dominating SHV extraction from advanced samples. As well as, as will be seen in Fig. S4 and S5, though the 2 strains ATCC®BAA-3272 and HFK3619 each produces different non-SHV-type β-lactamases, solely SHV was detected, indicating that the SHV-specific MEI-GP successfully distinguished SHV from different β-lactamase households, which was attributed to the rigorous screening to establish the enzyme family-specific epitope.
Mass spectra of SHV after extracted by MEI-GP from three strains of Gram-negative micro organism, together with A–C ATCC®25922 (Escherichia coli, doesn’t produce SHV), D HFK417 (Klebsiella pneumoniae, maintained by our laboratory, remoted from a medical specimen, produces SHV-1), and E ATCC®700603 (Okay. pneumonia, produces SHV-18), utilizing MNI-GP extraction and bacterial crackate resolution direct detection for comparisons. The spectrum was acquired from just one sediment-rich place with 40 laser photographs beneath a laser frequency of 40%⋅60 Hz
That is the primary examine to visually current bacterial resistance mechanism in a given mass spectrum, which mixed the specificity of epitope imprinting (SHV family-specific) with the sensitivity of mass spectrometry. The restrict of detection (LOD) of our method for SHV was ≤ 2 μg/mL, whereas the LOD for micro organism extraction was ≤ 15 mg, and the equal OD600 was ≤ 6.28 (micro organism dispersed in 1 mL of sterile water). The method of figuring out the LOD is described in Supplementary Data. By following this technique, different β-lactamase households can be particularly detected. In accordance with the molecular weight displayed in mass spectra, the form of β-lactamase and its related hydrolysis profile on β-lactams will be simply recognized. Primarily based on this, an preliminary drug choice scheme will be rapidly formulated for antimicrobial remedy. Utilizing this enzyme detection technique, from protein extraction to treatment steerage reporting, the imply time to detection (MTTD) was lower than 2 h.
Notably, realizing particular detection of SHV was not our final purpose, whereas that additional utility of the proposed technique to all the drug resistance-related proteins is the true function. Primarily based on this examine, following research may give attention to making use of a collection of enzyme family-specific supplies to assemble mass spectra libraries on AMR-related proteins, which may have nice implications for exactly greedy the enzyme epidemiological traits and rapidly formulating a complete drug choice scheme, thus successfully shortening the empirical treatment interval in medical medication.
Comparability with different drug resistance detection strategies
(i) In contrast with essentially the most generally used phenotype-based technique, antimicrobial susceptibility testing (AST) (MTTD: a minimum of 16–20 h) [38, 39], our enzyme detection technique avoids using antibiotics, and it takes not more than 2 h to acquire an preliminary AMR info on β-lactams, thus shortening the empirical treatment interval. However, as a golden customary of AMR detection, the AST technique supplies a variety of AMR info on a number of antibiotics. Subsequently, at the moment, our method may function a supplementary instrument for AST, by rapidly formulating an preliminary drug choice scheme for antimicrobial remedy, and quickly explaining complicated drug-resistant profiles. Nonetheless, sooner or later, by making use of a collection of enzyme family-specific supplies to assemble mass spectra libraries on AMR-related proteins, our proposed method might unseat AST dominance.
(ii) Hydrolysis-based strategies have been developed lately that depend on MS detection. Not like our enzyme detection technique, they’ll solely not directly detect β-lactamases by detecting the hydrolysates of β-lactams after an incubation of a number of hours or longer with micro organism [40]. Thus, these strategies are laborious and time-consuming, and non-specific to focus on enzymes. Conversely, by straight detecting β-lactamases, our method quickly explains the bacterial AMR profile on a variety of β-lactams at a time, thus rendering the method extra environment friendly.
(iii) Fingerprint-based strategies had been additionally developed within the final decade. Earlier than evaluation, a bacterial protein profile is obtained by MALDI-TOF MS with an m/z vary of 20,000–45,000 Da, which incorporates all the mass peaks of β-lactamases (29,000–39,000 Da). Nonetheless, trying to find goal enzyme peak from loads of peaks is a fairly sophisticated work, as a result of the enzyme sign is strongly interfered by matrix proteins, inflicting goal sign to be depressed and even disappear [41]. Compared, our enzyme-specific detection technique successfully eliminates matrix interferences, enabling β-lactamase peak to be clearly offered in mass spectra, and subsequently has a broad utility prospect.
(iv) As an rising gene modifying method, the CRISPR-Cas system has acquired widespread consideration in varied analysis fields, and has been utilized to AMR detection. Peng et al. designed a recombinase polymerase amplification-based CRISPR-Cas12a assay [42], by mixing Cas12a-crRNA (sgRNA) with the amplified fluorophore-labeled goal DNA fragments (AMR-related) to activate double-stranded breaks at a particular web site, and the discharge of fluorescence indicators indicated that the check pathogen was drug resistant. Equally, Allan-Blitz et al used an isothermal amplification-based CRISPR-Cas13a system to establish Neisseria gonorrhoeae through the porA gene and predict ciprofloxacin resistance by gyrA gene [43]. Clearly, the CRISPR-Cas system and our proposed method are each based mostly on the popularity of a particular fragment; the CRISPR-Cas system acknowledges a specific part (20–30 nucleotides) on a goal single-stranded DNA sequence, and our method acknowledges a particular epitope (9–15 amino acids) on a goal protein sequence. Nonetheless, in contrast with the current CRISPR-Cas-based method that’s extremely reliant on time-consuming DNA extraction, purification, and amplification, our method is straightforward to carry out in a short while, and the applying of MS detection will increase the strategy sensitivity.
(v) In contrast with different common gene-based methods, equivalent to oligonucleotide probe recognition, goal gene amplification, and whole-genome sequencing (MTTD: normally about 8 h) [14,15,16, 44], our enzyme detection technique can visually current bacterial resistance mechanism to researchers, and using imprinted materials (just like the probe in gene testing) endows our method with good specificity for goal enzymes, and subsequently is strongly complementary to the present gene testing. Furthermore, the technique of focusing on an enzyme household because the detection unit relatively than a single protein additional advances the work effectivity, rendering our method extra appropriate for medical utility.