HNRNPF interacts with Mir100hg and promotes its translocation from the nucleus to the cytoplasm in CSCs
Our earlier examine revealed that exosomes from LLC-SD improve the metastatic colonization functionality of LLC each in vitro and in vivo [20]. Right here, we noticed increased expression of Mir100hg in LLC-SD in comparison with LLC cells (Fig. S1A). Flattening Mir100hg in LLC-SD cells resulted in decreased ranges of Mir100hg within the derived exosomes (LLC-SD-Exo) (Fig. S1B, C). LLC-SD-Exo was characterised by way of Western blot, NTA, and electron microscopy (Fig. S1D–F). After co-culturing LLC-SD-Exo with LLC cells, the exosomes from the Mir100hg knockdown group considerably misplaced their means to advertise migration and invasion (Fig. S1G). Utilizing the mouse tail vein lung metastasis mannequin, the LLC-SD-sh-Mir100hg-Exo group exhibited fewer metastases within the lungs, coronary heart, and chest wall, and fewer weight reduction in comparison with the management group (Fig. 1A, B; S1H–J). These outcomes counsel that Mir100hg performs a important position within the oncogenic impact of LLC-SD-Exo.
HNRNPF interacts with Mir100hg and promotes its localization throughout the cytoplasm. A Schematic diagram of exosome-based animal experiments; B Fluorescence imaging of mouse lung tumor (inexperienced) and hematoxylin and eosin (HE) stained lung part; C Nucleoplasmic RNA isolation from LLC and LLC-SD cells; D Fluorescence in situ hybridization (FISH) of Mir100hg in LLC and LLC-SD cells; E RNA pulldown assay for Mir100hg; F Venn diagram of RNA pulldown outcomes; G Desk of knowledge associated to HNRNP household protein profiles in RNA-pulldown outcomes; H Nuclear and cytoplasmic RNA isolation from LLC-SD after flattening HNRNP household proteins; I FISH evaluation of Mir100hg; J RNA immunoprecipitation (RIP) for Mir100hg and HNRNPF; Ok RNA pulldown for Mir100hg and HNRNPF; L Fluorescence co-localization of Mir100hg and HNRNPF. M Schematic illustration of direct binding of Mir100hg to HNRNPF for nucleoplasmic translocation. (*p < 0.05, **p < 0.01, ***p < 0.001)
A number of research have demonstrated that Mir100hg is primarily localized within the nucleus, regulating transcription by both in a cis- or trans- method [21,22,23]. Intriguingly, Mir100hg displayed distinct localization in LLC-SD and LLC cells, with primarily cytoplasmic localization in LLC-SD and nuclear localization in LLC (Fig. 1C, D). To research the components contributing to the distinct nucleoplasmic localization of Mir100hg, RNA pull-down assay (RNA-PULLDOWN) was performed (Fig. 1E). Forty-five proteins sure to Mir100hg had been recognized, primarily concerned in localization and transporter pathyways (Fig. 1E, F, Fig. S2A, B; Supplementary Desk 1). Many of those proteins belong to the hnRNP household (Fig. 1G). Evaluation of The Most cancers Genome Atlas-Lung Adenocarcinoma (TCGA-LUAD) database revealed considerably increased ranges of those hnRNP proteins in lung adenocarcinoma sufferers in comparison with the non-tumor group (Fig. S2C). Excessive HNRNPF and HNRNPA2B1 expression correlated with decreased general survival (Fig. S2D).
To establish whether or not hnRNP household proteins mediate the nucleoplasmic translocation of Mir100hg, we silenced HNRNPs in LLC-SD cells utilizing small interfering RNA (siRNA) (Fig. S2E) and evaluated adjustments in Mir100hg localization. Silencing HNRNPF considerably altered Mir100hg localization, rising nuclear and lowering cytoplasmic ranges (Fig. 1H), confirmed by FISH outcomes (Fig. 1I). Evaluation of the mass spectrometry outcomes of HNRNPF in MIR100HG-pulldown (Fig. S2F), coupled with RNA immunoprecipitation (RIP), RNA-pulldown assays, Immunofluorescence co-localization and molecular docking simulations, confirmed a direct interplay between HNRNPF and Mir100hg (Fig. 1J–L; Fig. S2G). The expected secondary construction of Mir100hg was used within the RNA pull-down assay (Fig. S2H–J). The outcomes confirmed excessive binding affinity between Mir100hg and HNRNPF within the S1 (1-1247nt) and S3 (2155-3022nt) fragments (Fig. 1Ok). Collectively, these findings point out that HNRNPF enhances the cytoplasmic localization of Mir100hg by direct interplay, presumably resulting in elevated Mir100hg ranges within the exosomes of lung most cancers stem cells, thereby enhancing the metastatic potential of non-stem most cancers cells.
HNRNPA2B1 enhances the metastatic potential of LLC by facilitating the incorporation of Mir100hg into exosomes
To verify the exosomal switch of Mir100hg from LLC-SD to LLC, a Cy5-Biotin-Mir100hg tracing experiment was performed (Fig. 2A). Reverse transcription polymerase chain response (RT-PCR) and confocal microscopy verified its transfection into LLC-SD cells (Fig. S3A, B). Following co-culture of LLC-SD-Exo with LLC cells, exogenous Mir100hg was noticed in LLC cytoplasm (Fig. 2B), with elevated expression of Mir100hg in LLC cells (Fig. 2C), indicating the infiltration of LLC-SD exosomal Mir100hg into LLC cells. HNRNP silencing, notably HNRNPA2B1, resulted in decreased Mir100hg ranges in exosomes, suggesting its position in regulating the entry of Mir100hg into exosomes (Fig. 2D). Mass spectrometry, molecular docking, RIP, and RNA-PULLDOWN confirmed the direct binding between HNRNPA2B1 and Mir100hg (Fig. 2E, F; Fig. S3C, D), with a robust affinity for S1 (1-1247nt) and S2 (1247-2155nt) fragments (Fig. 2F). FISH and immunofluorescence assays confirmed colocalization of Mir100hg and HNRNPA2B1 in LLC-SD cells (Fig. S3E). Silencing HNRNPA2B1 in LLC-SD cells considerably decreased the extent of Mir100hg in exosomes (Fig. 2G; Fig. S3F, G). Co-culturing of LLC cells with exosomes from sh-HNRNPA2B1-LLC-SD and sh-NC-LLC-SD confirmed that HNRNPA2B1 silencing in LLC-SD cells decreased Mir100hg ranges in recipient LLC cells (Fig. 2H) and considerably diminished their migration and invasion functionality in vitro (Fig. 2I, J). Moreover, concomittent silencing of HNRNPA2B1 and HNRNPF in LLC-SD cells considerably decreased the migration and invasion of co-cultured LLC cells (Fig. S3H). These findings point out that the pro-metastatic impact of LLC-SD-Mir100hg on LLCs depends on HNRNPA2B1-mediated entry of Mir100hg into the exsomes.
HNRNPA2B1 enhances the metastatic potential of LLC by facilitating the incorporation of Mir100hg into exosomes. A Schematic diagram of exosomal Mir100hg tracing experiment; B Co-localization of Mir100hg and exosomes in LLC cells; C Bar graph displaying differential expression of Mir100hg in LLC cells post-exosome incubation; D Mir100hg expression in LLC-SD exosomes following HNRNP household protein knockdown; E RNA immunoprecipitation (RIP) for Mir100hg and HNRNPA2B1; F RNA pulldown for Mir100hg and HNRNPA2B1; G Mir100hg expression in LLC-SD cells following HNRNPA2B1 knockdown; H Mir100hg expression in LLC cells co-cultured with exosomes from HNRNPA2B1-knockdown LLC-SD cells; I, J Transwell for LLC cells post-co-culture with LLC-SD exosomes and the counting chart. (*p < 0.05, **p < 0.01, ***p < 0.001)
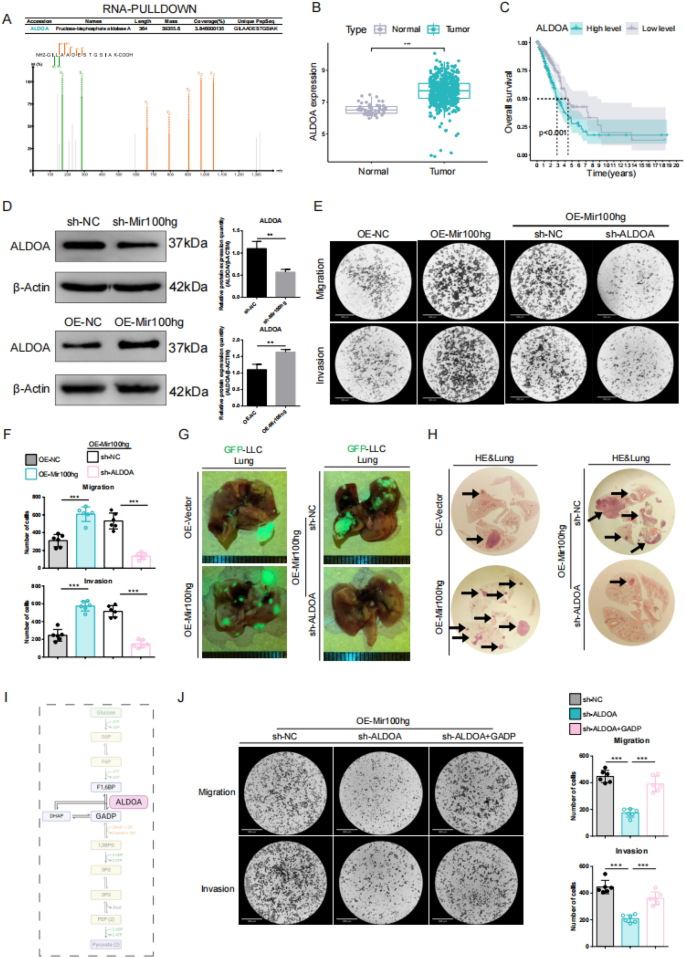
Mir100hg promotes the metastasis of lung most cancers cells in vitro and in vivo by up-regulating ALDOA expression
Most cancers growth and development contain metabolic shifts in cells to assist its speedy progress [24]. To establish key metabolic regulators downstream of Mir100hg, we carried out RNA-PULLDOWN coupled with mass spectrometry evaluation, which highlighted aldolase A (ALDOA) as the one glycolysis-related protein amongst Mir100hg-interacting proteins (Fig. 3A). ALDOA capabilities as a rate-limiting enzyme in glycolysis, and our evaluation of TCGA database revealed its vital overexpression in lung adenocarcinomas in comparison with non-tumor tissues, with excessive expression correlating with decreased affected person survival (Fig. 3B, C). These findings led us to hypothesize that Mir100hg promotes lung most cancers metastasis by upregulating ALDOA, thereby enhancing glycolysis. We demonstrated that ALDOA expression is positively correlated with Mir100hg expression (Fig. 3D). Overexpression of Mir100hg (OE-Mir100hg) enhanced the migration and invasion means of lung most cancers cells, whereas knockdown of ALDOA in OE-Mir100hg-LLC considerably reversed this phenotype (Fig. 3E, F). The mouse tail vein mannequin and hematoxylin and eosin(HE) staining outcomes additionally confirmed the same development (Fig. 3G, H). These outcomes counsel that Mir100hg enhances the metastatic means of lung most cancers cells in an ALDOA dependent method. ALDOA is well-known for its position in catalyzing the conversion of Fructose-1,6-bis-phosphate (FBP) into Dihydroxyacetone Phosphate (DHAP) and Glyceraldehyde-3-phosphate (GADP) in glycolysis (Fig. 3I). To research whether or not Mir100hg involving ALDOA impacts downstream metabolic processes, GADP was added to ALDOA knockdown cells. We noticed that GADP might restore the decreased migration and invasion skills brought on by ALDOA knockdown (Fig. 3J). These outcomes point out that Mir100hg enhances the migration and invasion means of lung most cancers cells, depending on the glycolysis course of involving ALDOA.
Mir100hg promotes the metastasis of lung most cancers cells in vitro and in vivo by up-regulating ALDOA expression. A Protein profile of ALDOA in RNA-PULLDOWN; B ALDOA expression variations in TCGA-LUAD database; C Line graph of the impact of ALDOA on general survival in TCGA-LUAD database; D Evaluation of ALDOA protein ranges in Mir100hg knockdown (sh-Mir100hg) and overexpression cells (OE-Mir100hg); E, F Transwell of the migration and invasion means of LLC cells and the counting chart; G Lung tumors in numerous mouse teams (inexperienced); H Hematoxylin and eosin (HE) stained lung sections (arrows point out tumor areas); I Schematic diagram of ALDOA involvement in glycolysis; J Transwell migration and invasion assays post-addition of GADP in ALDOA knockdown cells and the counting chart. (*p < 0.05, **p < 0.01, ***p < 0.001)
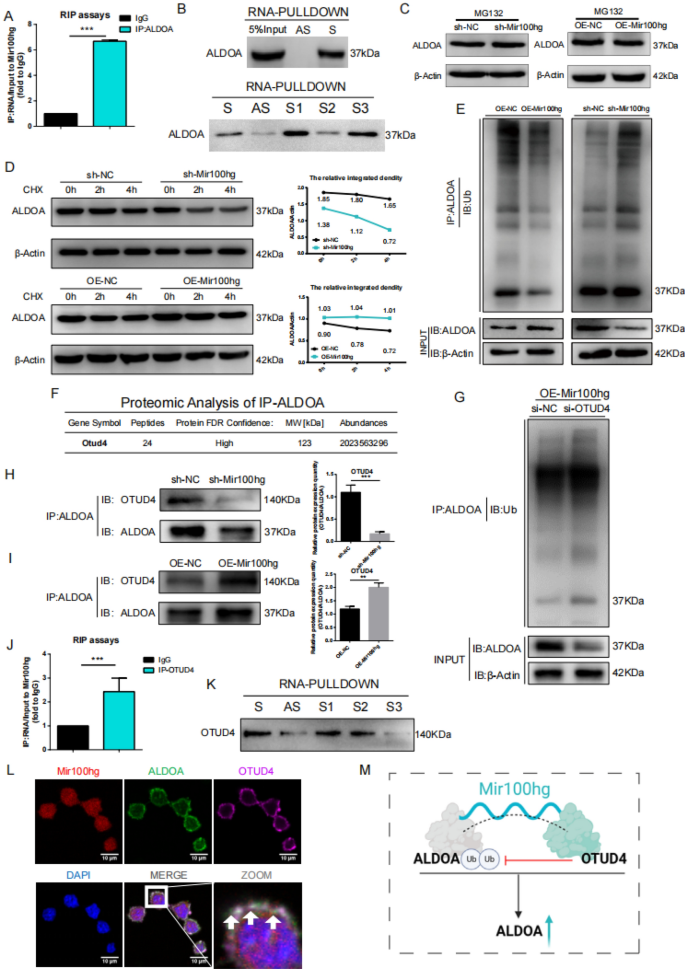
Mir100hg positively regulates ALDOA mRNA and protein ranges by CeRNA and OTUD4-deubiquitination
It’s well-established that lncRNAs can operate as scaffolds, influencing the ubiquitination of the sure proteins [25,26,27,28]. Molecular docking, RIP, and RNA-PULLDOWN (Fig. 4A, B; Fig. S4A) confirmed the direct binding between ALDOA and Mir100hg, with the S1/S3 fragment displaying stronger interplay. Using MG132, a proteasome inhibitor, can inhibit the constructive regulation of ALDOA by Mir 100hg, suggesting that Mir100hg might regulate ALDOA trough proteasomal degradation (Fig. 4C). To additional validate the position of Mir100hg in post-translational regulation of ALDOA degradation, cells had been handled with the protein synthesis inhibitor cycloheximide (CHX). Outcomes confirmed a sooner lower in ALDOA protein ranges in Mir100hg silencing (sh-Mir100hg) cells than in controls, whereas Mir100hg overexpression (OE-Mir100hg) delayed ALDOA degradation following CHX therapy (Fig. 4D). We subsequently investigated whether or not ubiquitination is concerned within the Mir100hg-dependent degradation of ALDOA. Immunoprecipitation evaluation revealed an inverse correlation between Mir100hg expression and ALDOA ubiquitination (Fig. 4E). These findings affirm that Mir100hg enhances ALDOA stability by the ubiquitin–proteasome system. To unravel the mechanism by which Mir100hg regulates the ubiquitin–proteasome degradation of ALDOA, we performed ALDOA pull-down assays and recognized OTUD4, a deubiquitinase (Fig. 4F; Supplementary Desk 2). The OTU household, as a significant deubiquitinating enzyme group, is essential for DNA restore, immune response, metabolic stability, and tumor growth [29,30,31,32,33]. Thus, we hypothesized that OTUD4 might promote ALDOA deubiquitination. Silencing OTUD4 in OE-Mir100hg cells considerably elevated ALDOA ubiquitination (Fig. 4G; Fig. S4B). We additional investigated whether or not Mir100hg regulates the interplay between ALDOA and OTUD4. Following Mir100hg silencing, there was a lower in OTUD4 protein co-precipitating with ALDOA in comparison with management cells (Fig. 4H), whereas Mir100hg-overexpression resulted in elevated co-precipitation of OTUD4 with ALDOA (Fig. 4I). Moreover, RIP and RNA-PULLDOWN experiments confirmed the sturdy and direct binding of the S1/S2 fragment of Mir100hg to OTUD4 (Fig. 4J, Ok). Immunofluorescence and FISH experiments revealed co-localization of Mir100hg, OTUD4, and ALDOA within the cytoplasm (Fig. 4L). Transwell assays demonstrated that OTUD4 knockdown in OE-100 cells considerably decreased lung most cancers cell migration and invasion in vitro (Fig. S4C, D). These findings counsel that Mir100hg recruits deubiquitinase OTUD4 to the ALDOA protein, thus impeding its degradation by way of the ubiquitin–proteasome system (Fig. 4M).
Mir100hg positively regulates ALDOA mRNA and protein ranges by CeRNA and OTUD4-deubiquitination. A RNA immunoprecipitation (RIP) for Mir100hg and ALDOA; B RNA pulldown for Mir100hg and ALDOA; C Evaluation of ALDOA protein ranges post-MG132 therapy in Mir100hg knockdown and overexpression cells; D Evaluation of ALDOA protein ranges and relative band depth quantification at indicated time factors post-CHX therapy in Mir100hg knockdown and overexpression cells; E Ubiquitination ranges of pulldown ALDOA protein analyzed by Western blot in Mir100hg knockdown and overexpression cells; F Protein spectrum outcomes for OTUD4; G Ubiquitination evaluation of ALDOA protein pulldown in OTUD4 knockdown cells by Western blot; H, I Evaluation of OTUD4 ranges following ALDOA protein pulldown in Mir100hg knockdown cells and Evaluation of OTUD4 ranges following ALDOA protein pulldown in Mir100hg knockdown cells; J, Ok RNA immunoprecipitation (RIP) and RNA-PULLDOWN for Mir100hg and OTUD4; L Fluorescence co-localization of Mir100hg with OTUD4 and ALDOA; M Schematic illustration of ALDOA ubiquitination inhibition by Mir100hg by OTUD4 (*p < 0.05, **p < 0.01, ***p < 0.001)
Our earlier examine demonstrated that Mir100hg targets two miRNAs, miR-15a-5p and miR-31-5p, by way of the aggressive endogenous RNA (CeRNA) regulation mechanism, thereby selling lung most cancers metastasis [20]. We hypothesized that Mir100hg might not solely straight affect ALDOA expression by protein stability but additionally modulates its mRNA ranges by way of miR-15a-5p and miR-31-5p. In OE-Mir100hg group, a lower in miR-15a-5p/miR-31-5p expression and elevated Aldoa mRNA ranges had been noticed (Fig. S4E), whereas knockdown of Mir100hg led to elevated miR-15a-5p/miR-31-5p expression and decreased Aldoa mRNA ranges (Fig. S4F). Moreover, simultaneous overexpression of miR-15a-5p and miR-31-5p in OE-Mir100hg-LLC cells considerably inhibited Aldoa mRNA ranges, whereas their suppression in sh-Mir100hg-LLC cells elevated Aldoa mRNA ranges (Fig. S4G, H). These findings affirm the molecular relationships of the transcriptional axis that entails the CeRNA community of Mir100hg, miR-15a-5p, miR-31-5p, and Aldoa. We then constructed dual-luciferase reporter techniques for each the wild-type and mutant plasmids of ALDOA (Fig. S4I). The discount in luminescence depth in cells transfected with Mir100hg-WT and Aldoa-WT plasmids signifies that these microRNAs can straight goal ALDOA. Importantly, the dearth of serious change in luminescence depth when mutations had been launched on the corresponding binding websites means that Mir100hg’s interplay with ALDOA is mediated by sequestering miR-15a-5p and miR-31-5p, stopping them from binding to ALDOA (Fig. S4J). These outcomes assist our speculation that Mir100hg binds to ALDOA and modulates its expression by sequestration of miR-15a-5p and miR-31-5p. To additional affirm the inhibitory impact of direct focusing on of ALDOA by miR-15a-5p and miR-31-5p on the metastatic potential of lung most cancers cells, we suppressed miR-15a-5p and miR-31-5p in wild-type LLC cells (Fig. S4K), which resulted in elevated ALDOA expression (Fig. S4L). Subsequent Transwell assays revealed that simultaneous suppression of miR-15a-5p and miR-31-5p considerably enhanced the migratory and invasive capabilities of LLC cells, whereas knockdown of ALDOA led to a noticeable discount in these capabilities (Fig. S4M). Importantly, we noticed that GADP might restore the decreased migration and invasion skills brought on by overexpressing miR-15a-5p and miR-31-5p (Fig. S4N). This implies that the downstream results of the CeRNA community regulated by Mir100hg are doubtless depending on the glycolytic course of involving ALDOA.
Taken collectively, these findings counsel that Mir100hg positively regulates ALDOA expression by CeRNA and OTUD4-mediated deubiquitination, which subsequently enhances the migration and invasion of lung most cancers cells.
Mir100hg facilitates lung most cancers cell metastasis by enhancing histone lactylation
Metabolomic evaluation demonstrated a major improve in intracellular lactate manufacturing following Mir100hg overexpression (Fig. 5A), suggesting that Mir100hg positively regulates lactate technology in lung most cancers cells. Moreover, using Fila-Nuc, a acknowledged lactate sensor [34], we noticed a major elevation in nuclear lactate ranges in OE-Mir100hg cells in comparison with the management group (Fig. S5A). Since lactate serves because the substrate for histone lactylation [15], we posit that Mir100hg might play a task within the regulation of histone lactylation in lung most cancers cells. Western blot and immunofluorescence assays revealed a major improve in histone lactylation ranges, notably at H3K9, H3K14, H3K18, and H4K16, with essentially the most pronounced improve noticed at H3K14 in OE-Mir100hg cells (Fig. 5B–D; Fig. S5B, C). Consequently, we targeted additional investigation on the impact of Mir100hg on H3K14 lactylation.
Mir100hg facilitates lung most cancers cell metastasis by enhancing lactylation. A Metabolomics heatmap; B World lactylation and H3K14la ranges detected by Western blot (WB); C, D Immunofluorescence detection of worldwide lactylation and H3K14la ranges; E Schematic diagram illustrating glycolytic steps affected by numerous medicine and inhibitors; F World lactylation and H3K14la ranges in OE-Mir100hg cells post-GNE therapy and subsequent NALA publicity; G Transwell of migration and invasion means of LLC cells after si-Ldha and the counting bar chart. H Transwell migration and invasion assays of LLC cells post-GNE and NALA and the counting bar chart (*p < 0.05, **p < 0.01, ***p < 0.001)
To validate the influence of glycolysis and histone lactylation on lung most cancers cell capabilities, we employed the glucose and glycolysis inhibitor 2-deoxy-d-glucose(2-DG), together with lactate dehydrogenase A inhibitor GNE140 (GNE) and sodium lactate (Nala) (Fig. 5E). The findings point out that glucose considerably enhanced the migration and invasion capabilities of lung most cancers cells (Fig. S5D), whereas 2-DG notably decreased their metastatic potential (Fig. S5E). These outcomes strongly counsel that glycolysis performs a vital position in augmenting the metastatic and invasive capacities of lung most cancers cells. Moreover, a major discount in international and H3K14 lactylation ranges was noticed in lung most cancers cells after GNE therapy (Fig. S5F), whereas Nala therapy led to will increase in these ranges (Fig. S5G). Moreover, in OE-Mir100hg cells, GNE decreased each international and H3K14 lactylation, which was successfully reversed by Nala (Fig. 5F). Transwell experiments demonstrated that lactylation inhibition induced by GNE attenuated lung most cancers cell migration and invasion in vitro (Fig. S5H), whereas Nala therapy elevated their metastatic potential (Fig. S5I). Notably, within the OE-Mir100hg group, GNE or si-Ldha decreased each migration and invasion, results that had been reversed by Nala (Fig. 5G, H).
Taken collectively, these findings counsel that Mir100hg enhances each international and H3K14 lactylation in lung most cancers cells, thereby contributing to lung most cancers metastasis.
H3K14 lactylation enhances the expression of genes related to lung most cancers metastasis
To research the position of H3K14 lactylation in lung most cancers, 4 experimental teams had been established: OE-NC, OE-Mir100hg, OE-Mir100hg with GNE therapy (GNE), and GNE therapy adopted by Nala restoration (GNE + NALA). Cleavage underneath targets and tagmentation (CUT&Tag) was employed to generate genome-wide H3K14la histone modification maps. The evaluation of genomic protection revealed elevated H3K14la modification within the OE-Mir100hg and GNE + NALA teams, primarily enriched on the promoter areas (Fig. 6A), with typical sharp peaks confined to transcription begin website (TSS) (Fig. 6B). RNA-Seq and CUT&Tag information confirmed the very best H3K14la enrichment at promoter areas within the high 25% expressed genes, with a progressive lower noticed in decrease quartiles, suggesting a constructive correlation between H3K14la and mRNA expression ranges (Fig. 6C). We analyzed Assay for Transposase-Accessible Chromatin sequencing (ATAC-Seq) information from GEO and located that genes with H3K14la enrichment in promoter exhibited increased chromatin accessibility in lung most cancers, indicating that H3K14la regulates transcription by establishing chromatin accessibility (Fig. 6D). We hypothesized that Mir100hg promotes lung most cancers cell metastasis by enhancing H3K14la-triggered gene transcription. RNA sequencing (RNA-Seq) and CUT&Tag information from the OE-Mir100hg and management teams revealed that upregulated genes in OE-Mir100hg group exhibited increased H3K14la accumulation at promoter areas (Fig. 6E). Gene set enrichment evaluation (GSEA) evaluation (Fig. 6F) revealed vital enrichment of tumor metastasis-related genes within the OE-Mir100hg group, with 63% of the upregulated genes displaying H3K14la enrichment (Fig. 6G). Kyoto Encyclopedia of Genes and Genomes (KEGG) evaluation of those genes urged a detailed affiliation with tumorigenesis and metastasis pathways (Fig. 6H). Moreover, we recognized 169 genes upregulated by Mir100hg overexpression and rescued by GNE, hypothesizing their regulation by the Mir100hg-H3K14la axis (Fig. 6I). Amongst these, 58% had been related to tumor metastasis (Fig. 6J), and 70% of those metastasis-associated genes confirmed vital H3K14la enrichment on the promoter area (Fig. 6Ok). Expression tendencies of a number of tumor metastasis-related genes elevated within the OE-Mir100hg group and decreased following GNE addition (Fig. 6L). PTGS2 was chosen for detailed evaluation, displaying vital will increase in expression and H3K14la peaks within the OE-Mir100hg group, which decreased following GNE addition (Fig. 6M). Equally, 145 genes regulated by the Mir100hg-H3K14la axis within the GNE and GNE + NALA teams. (Fig. S6A–E). These adjustments within the expression of tumor metastasis-associated genes and the H3K14la peaks of their promoters had been extremely correlated between the OE-Mir100hg group and the opposite teams (Fig. 6N). These findings counsel that Mir100hg enhances the metastatic potential of lung most cancers cells by facilitating the transcription of tumor metastasis-related genes by way of H3K14la.
Multi-omics evaluation reveals the involvement of the Mir100hg-H3K14la axis within the transcription of tumor metastasis-related genes. A Bar graph displaying the variety of H3K14la peaks enriched throughout 4 teams; B Heatmap of H3K14la enrichment in TSS areas throughout 4 teams; C Enrichment of H3K14la in TSS areas of genes exhibiting different expression ranges in RNA-Seq; D Differential chromatin opening in H3K14la-enriched genes inside TSS areas; E Differential heatmap in RNA-Seq of upregulated genes in OE-Mir100hg and corresponding TSS area heatmap in CUT&Tag; F GSEA evaluation indicating enrichment of differentially expressed genes in OE-Mir100hg inside tumor metastasis-related gene set; G Pie chart of the share of H3K14la-enriched genes amongst upregulated genes in OE-Mir100hg group; H KEGG evaluation of pathways related to H3K14LA-enriched upregulated genes; I–L Show of genes rescued post-addition of GNE in OE-Mir100hg group; M Ptgs2 peaks recognized in RNA-Seq and CUT&Tag; N Heatmap of TSS area enrichment for tumor metastasis-related genes in CUT&Tag and log fold change (logFC) heatmap in RNA-Seq (*p < 0.05, **p < 0.01, ***p < 0.001)
To establish the position of the Mir100hg-H3K14la axis in lung most cancers metastasis, 5 key genes had been recognized by CUT&Tag, RNA-Seq, RT-PCR and prognosis evaluation (Fig. 7A). Evaluation of the TCGA-LUAD database confirmed the excessive expression of those 5 genes in lung adenocarcinoma in comparison with non-cancerous tissues (Fig. 7B). In comparison with the management group, overexpression of Mir100hg considerably elevated H3K14 modifications close to the TSS and expression ranges of those 5 genes. Beneath Mir100hg overexpression, GNE decreased the H3K14 modifications and expression ranges of those genes, whereas NALA considerably restore them (Fig. 7C). Moreover, survival evaluation urged that top expression of those genes correlated with considerably decrease general survival in lung adenocarcinoma sufferers. Lastly, following siRNA-mediated knockdown of those 5 genes, transwell assays had been carried out. As proven in Fig. 7D, the knockdown of the 5 genes considerably inhibited lung most cancers cell migration.
The Mir100hg-H3K14 lactylation axis enhances the expression of genes related to tumor metastasis. A Movement chart for gene screening course of; B Boxplot of LMAN1, OSTC, P4HA1, TEX30, and TMEM65 gene expression variations in TCGA-LUAD; C Lman1/Ostc/P4ha1/Tex30/Tmem65 in CUT&Tag peak determine show, mRNA expression adjustments and survival evaluation; D Transwell for cell migration and invasion post-knockdown of LMAN1, OSTC, P4HA1, TEX30, and TMEM65 and the counting bar chart (*p < 0.05, **p < 0.01, ***p < 0.001)
Collectively, our findings counsel that Lman1, Ostc, P4ha1, Tex30, and Tmem65 are downstream genes selling tumor metastasis, influenced by the Mir100hg-H3K14la regulatory axis in lung most cancers.