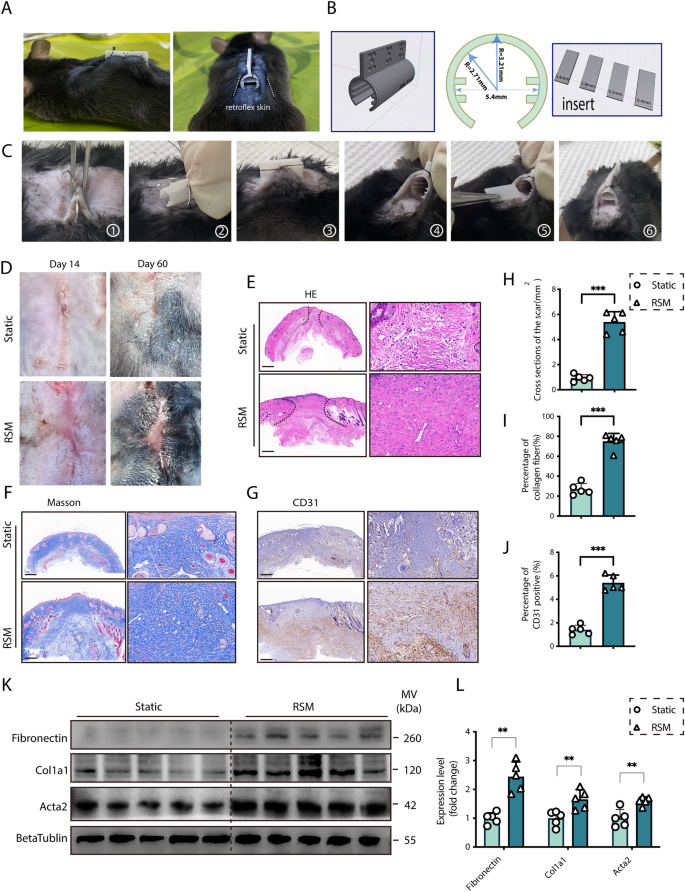
Experimental design
The aim of this research is to make the most of rigidity to induce and set up a steady and reproducible mouse scar mannequin, which can be utilized to conduct in-depth analysis on the pathogenesis of tension-induced PS On this research, we recognized and validated the important thing function of Sele within the formation of tension-induced PS. Primarily based on this discovery, we designed possible siRNA lipid nanoparticles (LNPs) concentrating on Sele to particularly silence Sele expression in endothelial cells, and verified the therapeutic impact of this technique in scar mannequin mice. The physicochemical properties of the focused siSele LNP have been characterised by measuring dimension, morphology, zeta potential, encapsulation effectivity (EE), and stability. Major vascular endothelial cells might be extracted from the pores and skin to evaluate the protection, uptake potential, and talent to focus on and silence genes in LNP. In accordance with the protocols authorized by the Institutional Animal Care and Use Committee of the Ninth Individuals’s Hospital affiliated with Shanghai Jiao Tong College Faculty of Drugs, the in vivo concentrating on potential and therapeutic impact might be evaluated in C57/BL6 mice. Mice might be randomly assigned to every research group. All experiments on this research might be carried out utilizing at the least 5 unbiased replicates.
Ethics assertion
Six to eight weeks previous Male C57/BL6 have been bought from Shanghai JieSiJie Laboratory Animal Co., Ltd. The research was carried out in compliance with moral pointers and was authorized by the ethics committee of Shanghai Ninth Individuals’s Hospital, affiliated with Shanghai Jiao Tong College Faculty of Drugs, underneath the approval quantity SH9H-2020-A314-1. Efforts have been made to attenuate the variety of animals used and alleviate any discomfort they may expertise. PS (keloid) tissue and regular pores and skin tissue have been each taken from the Division of Plastic Surgical procedure and Dermatology of Shanghai Ninth Individuals’s Hospital, affiliated with Shanghai Jiao Tong College Faculty of Drugs, together with 5 instances of keloid tissue (Desk 1) and 5 instances of regular pores and skin tissue as a management group. The medical research was authorized by the moral overview board of, underneath the approval quantity SH9 H-2024-TK400-1.
Retroflex scar mannequin (RSM) in mice
In our research, a linear incision with a size of 1.5 cm was made on the dorsal facet of mice, and the wound was sutured with 5–0 suture. Take away the suture on the third day to make sure that the linear wound doesn’t rupture. After eradicating the stitches, retroflex the pores and skin 1 cm away from either side of the linear wound alongside the curved tensile gadget, and use 3–0 suture on the prime of the tensile gadget to repair the pores and skin on either side collectively by means of small holes, forming a cylinder composed of the pores and skin on either side curled as much as preserve the pores and skin beneath the wound tight. The design drawing of the tensile gadget producer is proven in Fig. 1B. The mildew producer is a 300-degree cylindrical construction made from Polyethylene (PE) materials, utilizing 3D printing expertise, with small holes on the prime for fixing the pores and skin on either side. The strain of the pores and skin mounted on the cylinder is adjusted in keeping with the width of the insert in the midst of the cylinder. Measure the gap of pores and skin extension on the wound web site on days 0, 3, 7, and 14 after modeling, and make sure that the pores and skin on the wound web site is at all times saved underneath excessive rigidity by well timed inserting spacers of various widths, the detailed calculation formulation are supplied in supplementary Textual content. Mice in static group underwent the identical linear incision and suture because the RSM mannequin. After suture elimination, the injuries have been allowed to heal naturally with none gadget intervention.
Building and characterization of a novel stretch-mediated mice RSM. A Facet and entrance views of the mice Retroflex scar mannequin (RSM). B Schematic of the RSM tensile gadget, highlighting the scale of cylindrical physique and insert. C The process of molding. D Gross morphology of scars in management (Static) and RSM teams on days 14 and 60 post-incision. Histological assessments at day 14: E H&E staining, F Masson’s trichrome, and G CD31 immunostaining scale bar = 200 μm. H–J Semi-quantitative evaluation of histology for management and RSM teams on day 14. Okay–L Western blot evaluation and semi-quantification of Fibronectin, Col1a1, and Acta2 at day 14 (n = 5. *p < 0.05, **p < 0.01, ***p < 0.001, ns = not important)
Extraction of main human dermal vascular endothelial cells and cell tradition
Contemporary human pores and skin tissue was sourced from cosmetic surgery, knowledgeable consent was obtained from the sufferers or their authorized representatives. Pores and skin tissue was minimize into small items (roughly 1–2 mm3). The tissue items are then digested utilizing a mix of collagenase I (biosharp, BS163), hyaluronidase (20426ES60) and trypsin–EDTA (Gibco, 25200056). The digested tissue is then gently triturated to launch cells, and the cell suspension is filtered by means of a 70 μm cell strainer (corning, CLS431751) to take away undigested tissue fragments. The cell suspension is centrifuged at 300g for 10 min, the supernatant is discarded, and the cell pellet is resuspended in endothelial cell development medium EGM-2 (Clonetics, CC-3162). Endothelial cell line HMEC-1 was obtained from Nationwide Assortment of Authenticated Cell Cultures, and cultured in endothelial cell development medium EGM-2 (Clonetics, CC-3162). Monocyte cell-line THP-1 was cultured in RPMI-1640 medium (SH30809.01, HyClone, USA) containing 10% FBS and 1% penicillin/streptomycin/fungizone (15240-062, Gibco, USA).
Monocyte-endothelial cell adhesion assay
Major endothelial cells have been digested from a static CMS gadget or a stretched CMS gadget and cultured in confocal dishes (FCFC020-10pcs, Beyotime). THP-1 cells have been stained with CellTrace (40717ES50, Yeasen) for 20 min earlier than coincubation with the endothelial cells for 30 min. Statement and counting of adherent THP-1 cells utilizing a microscope (Zeiss, Germany) and ImageJ software program.
CRISPR/Cas9 mediated Sele knockout
The deletion of the Sele gene in HMEC-1 cell strains was carried out. (Cas9X, China). In short, the gene sequence of Sele was retrieved from the NCBI database, and applicable sgRNA was designed and used to assemble plasmids. Utilizing CRISPR/Cas9 gene enhancing expertise, the Cas9 and sgRNA plasmids have been electroporated into the cells. CRISPR/Cas9-mediated Sele endothelial cells have been then screened utilizing PCR and confirmed to be deleted.
Histology and immunohistochemistry
Lower off the scar tissue and rapidly immerse it in 4% paraformaldehyde for twenty-four h. Dehydrate the scar tissue and embed it in avidin, then slice it and use H&E, and Masson’s tri-cochrome staining. For immunohistochemistry evaluation, tissue sections have been handled with applicable main antibodies adopted by incubation with HRP-conjugated secondary antibodies. The three,3′-diaminobenzidine (DAB) substrate was used for direct visualization of the distribution of goal proteins.
Western blot and co-immunoprecipitation (CO-IP)
Protein samples from cells and tissue have been extracted utilizing the Radio Immunoprecipitation Assay (PC101, Epizyme) and quantified utilizing the BCA Protein Assay (ZJ102, Epizyme). After denaturing and separating on 7.5% or 12.5% Sodium Dodecyl Sulfate–Polyacrylamide Gels (SDS-PAGE), the protein on SDS-gels have been transferred to a PVDF membrane (IPVH00010, Milipore), and incubating with the first antibodies as follows: Sele (1:1000, Santa Cruz, sc-137054), Atpb (1:1000, P06576, Abmart), Gapdh (1:50,000, 60,004-1-Ig, Proteintech), Col1a1(1:5000, 67288-1-Ig, Proteintech), Acta2(1:20,000, 67735-1-Ig, Proteintech), PSGL-1 (1:1000, T58338, Abmart), CD44(1:1000, T55122, Abmart), CD43(1:1000, PU202528, Abmart). Secondary antibodies have been additional used as follows: HRP-labeled Goat Anti-Rabbit IgG(H+L) (1:1000, A0208, Beyotime), HRP-labeled Goat Anti-Mouse IgG(H+L) (1:1000, A0216, Beyotime). For co-IP, protein pattern was lysed by lysis buffer for IP (P0013, Beyotime). Earlier than the co-IP, pre-incubated lysate with Protein A/G Magnetic Beads (HY-K0202, MedChemExpress) with rotator at 4 °C for two h for eradicating non-specific proteins certain to the beads. Subsequently, add diluted antibody (Ab) to the lysate above right into a 1.5 mL tube for in a single day at 4 °C, Mouse IgG (1ug for IP, B900620, Proteintech) have been used as a detrimental management. Add the Protein A/G Magnetic Beads into antigen–antibody (Ag-Ab) advanced and rotate tube for two h at 4 °C. Accumulate the protein–protein complexes, they have been later subjected to Western blot for the additional experiment.
RT-qPCR
Trizol reagent (Invitrogen, United States) was employed for complete RNA extraction. The extracted RNA samples have been then reverse-transcribed into cDNA utilizing the PrimeScriptTM RT reagent Equipment (TAKARA, Japan). RT-qPCR was carried out on a Gentle Cycler thermal cycler system (Bio-Rad, United States) utilizing SYBR® Premix Ex Taq™ II (TAKARA, Japan). For mRNA normalization, GPADH was used as the interior management. The relative expression was in comparison with the management group. The sequences of the primers used within the current research are proven in Desk 2.
Transcriptome sequencing
Second-generation sequencing was carried out on the PS tissue from the RSM group and regular scar tissue from the management group. Sequencing was carried out utilizing the Illumina NovaSeq™ 6000 platform with paired-end (PE) 150 bp sequencing mode. The outcomes have been analyzed through R (v4.3.1) software program.
Immunofluorescence
After administering anesthesia to the mice, the scar tissue was extracted with 4% paraformaldehyde. The scar tissue was dehydrated utilizing sucrose and embedded in OCT compound (Sakura, Japan) earlier than quickly frozen at − 20 °C. Subsequently, the scars have been coronally sliced into sections with a thickness of 30 μm. The scar sections underwent a triple wash with PBST and have been incubated in a single day at 4 °C with particular antibodies CD31(1:100, 11265-1-AP, Proteintech), CD11b (1:100, sc-52600, santa Cruz) and Sele (1:100, TD6914, Abmart). Following one other spherical of triple washes with PBST, the sections have been uncovered to completely different secondary antibodies (Invitrogen, United States) for 1 h at room temperature. After a closing triple wash with PBST, the pores and skin sections have been mounted utilizing a DAPI-containing mounting medium (southenbiotech, United States). Digital pathology syestem was used to watch and seize photographs of sections.
Single-cell RNA sequence information evaluation
The one-cell RNA-seq information for this research have been obtained from GSE181316 and GSE156326, together with human regular scar, human Keloid and human HS. High quality management and cell annotation have been carried out in keeping with a previous research. R bundle Seurat (v4.1.3) was used for information integration, cell filtration, normalization, clustering, and Uniform Manifold Approximation and Projection (UMAP) dimensional discount. Cell development genes have been outlined primarily based on DEGs amongst Seurat clusters. Inference and evaluation of cell–cell communication have been used through R bundle CellChat to v2.
Mechanical stretch gadgets
Utilizing FX-5000T ™ The Flexcell Rigidity Plus system (Flexcell Worldwide Company, Hillsborough, NC, USA) applies cyclic mechanical stretching (CMS) to endothelial cells. Endothelial cells have been cultured at a charge of two * 105 cells/mL on a 6-well BioFlex tradition (Flexcell Worldwide Company, Hillsborough, NC, USA) with a versatile silicon membrane backside. CMS is utilized to the stretching group in a sinusoidal mode with an amplitude of 0%–20% at 0.5 Hz. Static cells cultured in the identical kind of plate however saved stationary have been incubated in the identical incubator.
Irritation array and ELISA array
Quantibody® Human Irritation Array 1 by RayBiotech (Norcross, GA, USA) was utilized to quantitatively detect the supernatant of static or stretched Endothelial-Monocytes co-culture system. It employs particular antibody pairs designed for this goal, the experimental protocol and information evaluation procedures really helpful by Ray Biotech have been strictly adopted. Cytokines displaying statistical significance (p < 0.05) throughout completely different experimental teams have been thought of statistically important. To facilitate information visualization and interpretation, a volcano plot was generated utilizing the R statistical software program model 4.3.1, permitting for the identification of developments and patterns within the information. To research the particular secretion pro-inflammation cytokines, ELISA have been employed. After numerous experimental interventions, tradition medium was harvested, adopted by centrifugation at 400×g for five min at 4 °C. The ensuing supernatants have been meticulously collected and subjected to quantification utilizing a Human IL-1α ELISA package (PI565, Beyotime), Human IL-6 ELISA package (ab178013, Abcam), and Human TNF-alpha ELISA Equipment (ab108908, Abcam).
Preparation of siRNA@LNPs
LNPs are ready by skinny filming-rehydration methodology from cationic lipids DOTAP (MCE, USA), DOPE (MCE, USA), ldl cholesterol (McLean, USA), and DSPE-PEG2000 (MCE, USA). Dissolve DOTAP, DOPE, ldl cholesterol, and DSPE-PEG2000 (so as of 5, 5, 0.5, and 0.11 mg) in chloroform (3 mL), and switch the lipid resolution to a spherical backside flask after ultrasonic therapy to totally dissolve it. Steam underneath vacuum situations for 30 min (75 r/min, room temperature) to permit the natural solvent to utterly evaporate and procure a homogeneous lipid movie. Hydrate with 3 mL of enzyme free water for 10 min (100 r/min) after which homogenize with ultrasound. Then, the siRNA resolution was combined with LNPs by electrostatic interplay at a weight ratio of 1:30, and concentrated by centrifugation in a Macrosep Advance filtration gadget (MAP001C37) (3000g/min, 4 °C). The molecular weight was minimize off to 10 kDa to take away the unsealed drug.
siRNA@LNPs stability evaluation and physicochemical characterization
To check the soundness of bare siRNA and siRNA@LNPs, incubating bare siRNA or siRNA@LNPs with RNase (10 μg/mL) at 37 °C for 0, 15, 30, 60, 120, or 240 min. Then gather LNPs by centrifuging at 12,000 rpm for 10 min. Then extracted siRNA in 0.5 M NaCl containing 0.1% SDS (58). Electrophoresis was carried out on 3% agarose gel injected by GelRed, after which imaging was carried out underneath ultraviolet (UV) gentle. The particle dimension of LNPs was measured earlier than and after stretching utilizing DLS (Malvern, UK). Transmission electron microscope (TEM) (Thermo Fisher Scientific, USA) was used to characterize the morphology of LNPs earlier than and after stretching. The dimensions and zeta potential of LNPs have been measured utilizing DLS.
Willpower of encapsulation effectivity
Measure the encapsulation effectivity (EE) in keeping with the directions of Quant-iT RiboGreen reagent. Put together LNP loaded with siRNA in keeping with the above methodology. Combine 3 μ l quantity of LNP resolution with 117 μL of 1 × TE (Tris EDTA) buffer or 2% Triton X-100, and vortex for two min. Dilute the free siRNA commonplace with a sequence of concentrations. Incubate NP samples and siRNA requirements with an equal quantity of 1:200 diluted RiboGreen reagent for five min. Measure fluorescence depth utilizing an enzyme-linked immunosorbent assay (excitation/emission, 480/520 nm; Tecan, Switzerland). Calculate the encapsulation effectivity (EE%) utilizing the next formulation:
$$ {textual content{EE}}left( % proper) = left( {{textual content{fluorescence}};{textual content{of}};{textual content{B}} – {textual content{fluorescence}};{textual content{of}};{textual content{A}}} proper)/left( {{textual content{fluorescence}};{textual content{of}};{textual content{B}}} proper) occasions {1}00% $$
Biodistribution research
Twenty-four hours after native injection of the cy5-siSele@LNPs on the wound web site, mice have been subjected to dwell imaging on the IVIS system (Perkin Elmer, USA). Instantly after imaging, the mice have been sacrificed and their hearts, liver, spleen, and lungs have been eliminated for imaging on the IVIS system. To find out cy5-siSele@LNPs Cell focused assortment, shredding, and digestion of scar tissue in PBS containing collagenase I (201.3U/ml; Yeason), 0.92 M Hepes (Beyotime), and DNase I (50.3U/ml; Sigma Aldrich) at 37 °C for 1 h. Filter the obtained digests utilizing a 70 μm cell filter and deal with with purple blood cell lysis buffer for five min. Then centrifuge the pattern at 400g, resuspend it in PBS containing 0.5% BSA, and filter it by means of a 40 μm cell filter. Subsequently, the pattern was incubated with antibodies concentrating on epithelial cells (EpCAM Alexa Fluor 647), immune cells (CD45 phycoerythrin), and endothelial cells (CD31 Shiny Purple 421) markers at 4 °C for 30 min. Utilizing circulate cytometry (BD Biosciences, USA) to find out numerous cell subtypes cy5-siSele@LNPs stage.
Statistical evaluation
All experimental information have been expressed because the imply ± commonplace deviation (imply ± SD) and analyzed utilizing GraphPad Prism 9.0 software program (La Jolla, CA, USA). A t-test was used for comparisons between two teams. For comparisons amongst a number of teams, one-way ANOVA adopted by Tukey’s a number of comparability check was carried out. All assessments have been two-tailed, with p < 0.05 indicating statistical significance.